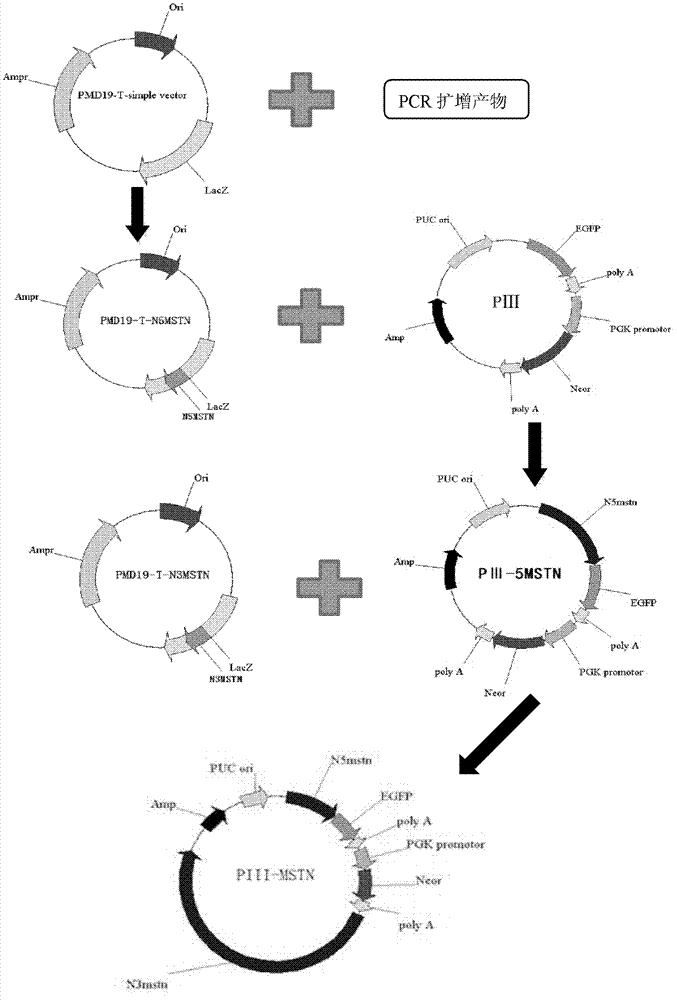
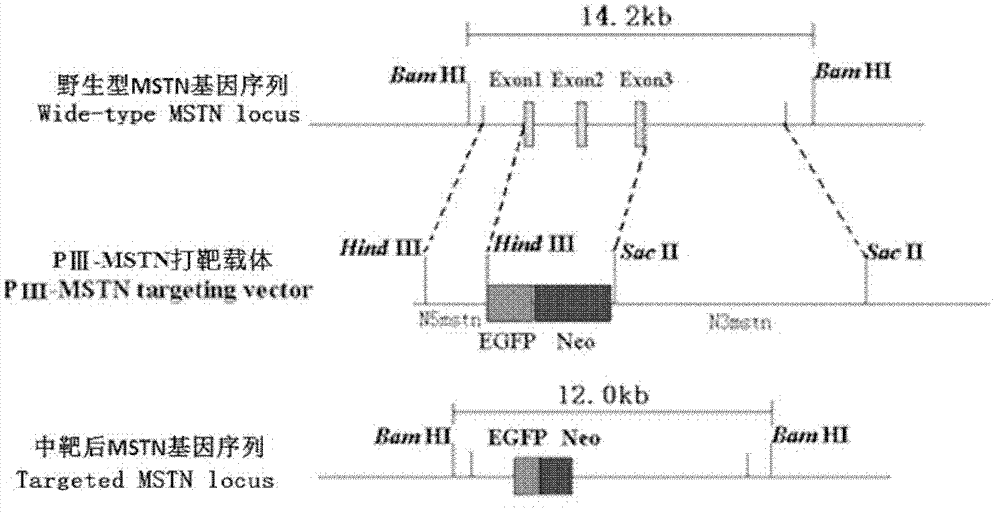
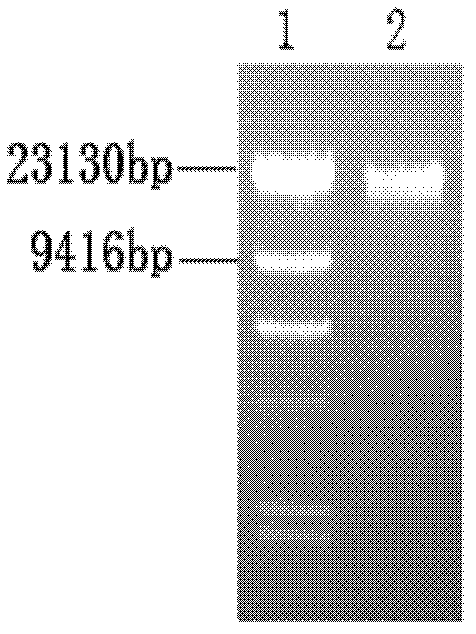
Targeting vector for knockout of bovine MSTN gene and application thereof
A technology for targeting vectors and genes, which is applied in the fields of molecular biology and biology to simplify the screening work
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The extraction of embodiment 1 bovine ear tissue genomic DNA
[0037] Weighed 500 mg of ear tissue from Wagyu cattle (from Siziwangqi Ranch in Inner Mongolia Autonomous Region) stored in -80°C ultra-low temperature freezer, and used Promega’s The Genomic DNA Purification Kit was used to extract genomic DNA from Wagyu ear tissue according to the DNA extraction method of rat tail tissue, and finally the DNA was redissolved in 100 μL sterile distilled water. Use an ultraviolet spectrophotometer to detect the concentration of the extracted DNA and the OD value at a wavelength of 340 nm, and take 1 μL for agarose gel electrophoresis to detect the integrity of the DNA. The results of electrophoresis showed that the complete Wagyu genomic DNA was extracted, see image 3 .
Embodiment 2
[0038] Example 2 Obtaining of MSTN gene homology arms pMD19-T-N5mstn and pMD19-T-N3mstn
[0039] According to the template sequence GenBank Accession No.NC_007300, the upstream and downstream primers ShortA (TTT AAGCTT TCATTTGATTAGACCTTGTGGCTCC) and ShortB (TTT AAGCTT GGTTTTAAAA TCAATACAATCTTTT), and the upper and lower primer LongA (GA TCCGCGG CATGGTAGTAGATCGCTGTGGGTGT) and LongB (AAT CCGCGG AGTAGAATGGTCTTGAATGGTTAGGA), the underlined part is the restriction site. The 5' and 3' homology arms of the MSTN gene of the promoter-trapping targeting vector were synthesized by PCR using the above-mentioned bovine ear tissue genomic DNA as a template.
[0040] All PCR reactions are 20 μL reaction system: 2 μL of upstream and downstream primers, 0.5 μL of template DNA, 2 μL of 10×Buffer, 25 mM MgCl 2 1.4 μL, 2.5 mM dNTPs 3 μL, EXTaq 0.2 μL, sterilized ddH 2 O 8.9 μL.
[0041] PCR reaction conditions for the 5' homology arm: denaturation at 95°C for 50 s, annealing at 60°C for...
Embodiment 3
[0043] Example 3 Enzyme digestion identification and sequencing analysis of the 5' homology arm pMD19-T-N5mstn
[0044] Take 1 μL of the 5' homology arm pMD19-T-N5mstn plasmid, and use restriction endonucleases HindIII and Bgl II to carry out enzyme digestion and identification, and their restriction sites are located at both ends and inside of the 5' homology arm, respectively. Preliminary detection of the correctness of the 5' homology arm obtained by PCR. All enzyme digestion systems are 10 μL: 1 μL of 5’ homology arm pMD19-T-N5mstn plasmid, 10×Buffer 1 μL, endonuclease 0.5 μL, sterilized ddH 2 O 7.5 μL. DNA sequencing analysis was carried out on the correct plasmid identified by enzyme digestion, and the results showed that the homology of the 5' homology arm with the bovine MSTN gene sequence reported by GenBank was 96%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com