Method for quickly building differential expression gene library
A technology of differentially expressed genes and libraries, applied in the biological field, can solve the problems of high cost, many operation steps, and the inability of ordinary laboratories to achieve the effects of reducing experimental costs, simplifying operating procedures, and reliable technical support
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] The preferred embodiments of the present invention will be described in detail below with reference to the accompanying drawings.
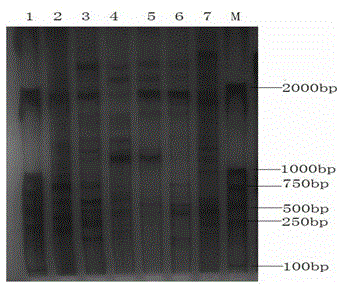
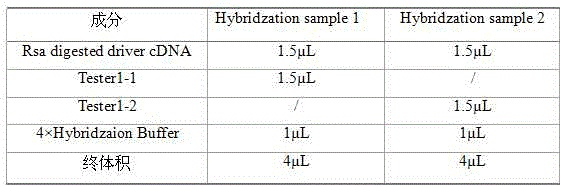
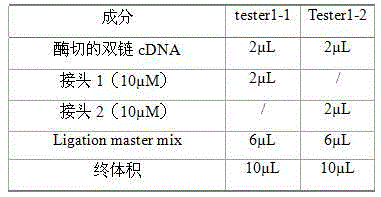
[0023] 1. Cloning-free suppression subtractive hybridization (ACSSH) reagents: random primers, 5×First-strand Buffer (first-strand buffer), ligation buffer, hybridization buffer, Taq polymerase buffer, Second-strand Enzyme Cocktail ( Second strand synthetase), 5×Second-strand Buffer (second strand buffer), dNTPs, ddH 2 O, AMV reverse transcriptase, T4 DNA polymerase, EDTA / Glycogen Mix (EDTA / sugar mixture), phenol:chloroform:isoamyl alcohol 25:24:1 (volume ratio), Rsa I enzyme, Rsa buffer, T4 DNA ligase, Tester1-1 (transcript 1), Tester1-2 (transcript 2), Adaptor1 (linker 1), Adaptor2 (linker 2), 5×Ligation Buffer (ligation buffer), Ligation master mix (ligation Reaction solution) and Taq DNA polymerase, the above reagents were purchased from Shanghai Sangon Bioengineering Technology Co., Ltd.
[0024] 2. Denaturing gradient gel (DGGE) rea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com