Method of measuring glycosyl in protein
A protein and protease technology, applied in the field of biological protein detection, can solve the problems of difficult to preserve the sugar chain structure, incomplete sugar cleavage, poor reproducibility, etc., and achieve the effect of complete sugar chain structure and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 3
[0127] 1. Experimental steps
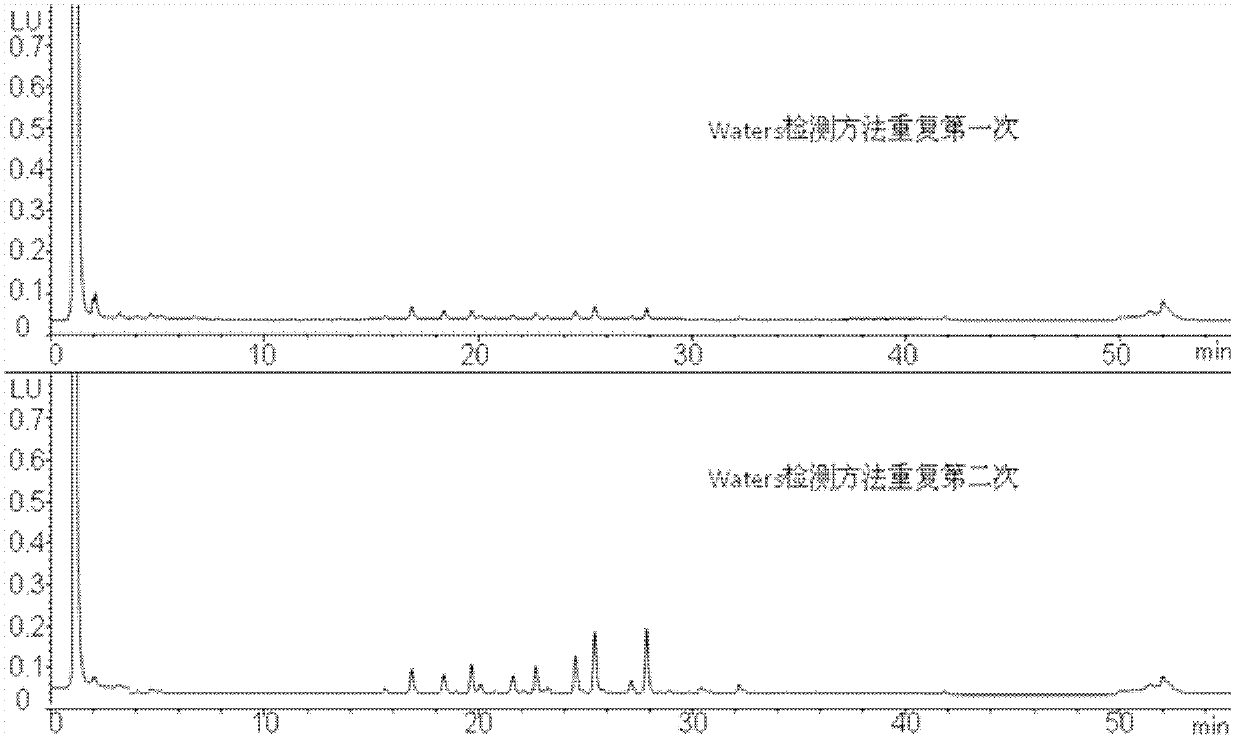
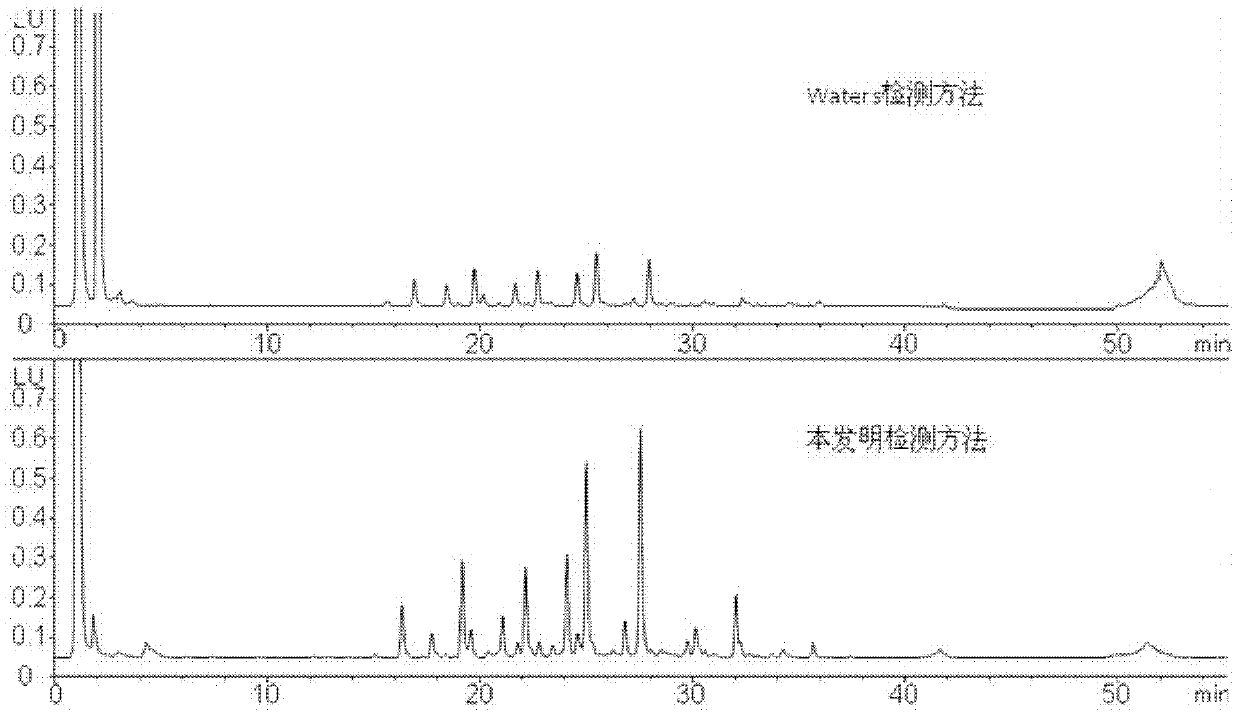
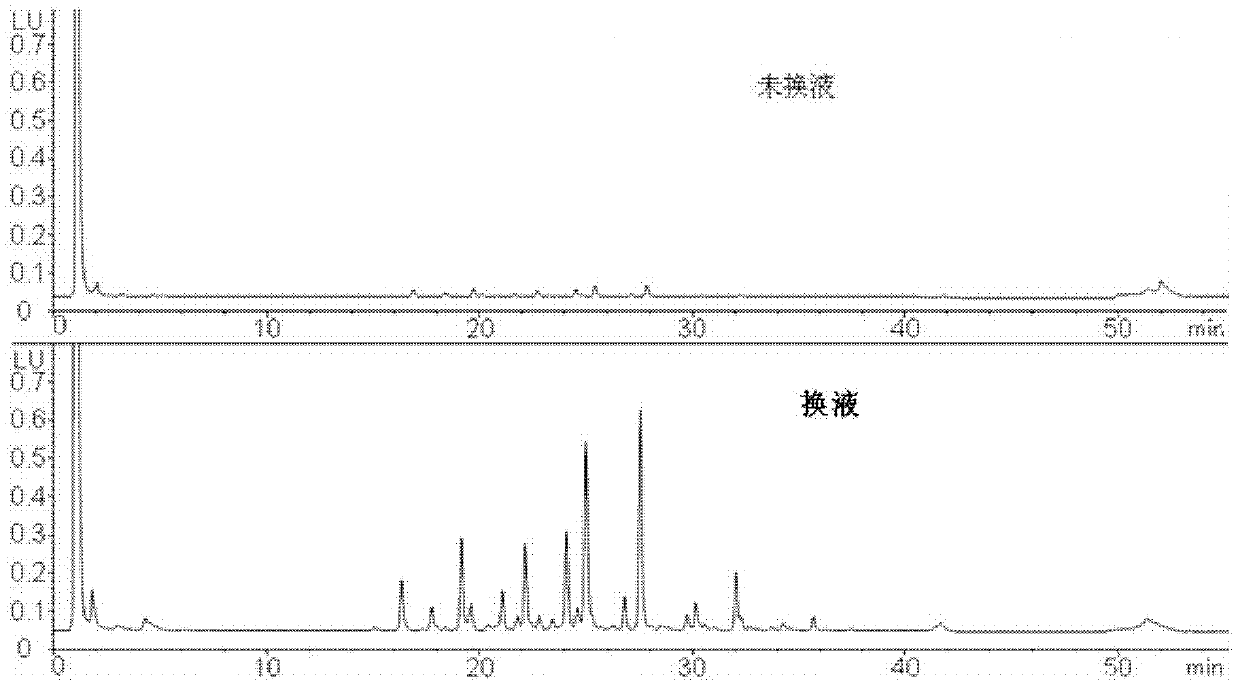
[0128] 1) Liquid exchange process
[0129] Take 1mg of protein (sequence 1) solution, put it into an ultrafiltration centrifuge tube, supplement 400μl with 50mmol / L ammonium bicarbonate buffer (pH=8.3), centrifuge at 10000r / min for 10min, then add 350μl 50mmol / L ammonium bicarbonate buffer (pH=8.3), centrifuge at 10000 r / min for 10 min, and repeat the operation more than 3 times. At this time, the sample volume is about 40 μl, and the concentration is about 25 mg / ml.
[0130] 2) Sample digestion and sugar cutting
[0131] Take 10 μl of the protein (sequence 1) solution after liquid exchange treatment into a clean 1.5ml EP tube, and do two parallel experiments. Add an equal volume of 8M guanidine hydrochloride solution to the samples, mix well, and denature the protein in a 65°C water bath for 30 minutes; take the denatured sample, add 1 μl of 1M DTT solution to make the final concentration of DTT 50 mM, and reduce the protein in a 40°C water ba...
Embodiment 4
[0145] The detection of embodiment tetrasialic acid
[0146] Sialic acid is located at the outermost side of the sugar chain, and it is easy to fall off during the analysis process. In order to better reflect the pretreatment process and detection method of the present invention, the structure of the sugar chain is not changed, especially according to Example 1 (sample 1- 3) and Example 3 (sample 4-6), the sugar chains obtained by enzymatic cleavage and then sugar cleavage were used for detection of sialic acid.
[0147] The inventors determined the content of sialic acid in protein according to the resorcinol method recorded in the 2010 edition of "Chinese Pharmacopoeia", the third appendix VIC sialic acid determination method.
[0148] Calculate the content of sugar chains according to the HPLC results obtained by the method for detecting sugar groups in proteins according to the present invention, and the mass spectrometer detection results know the number of sialic acid in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com