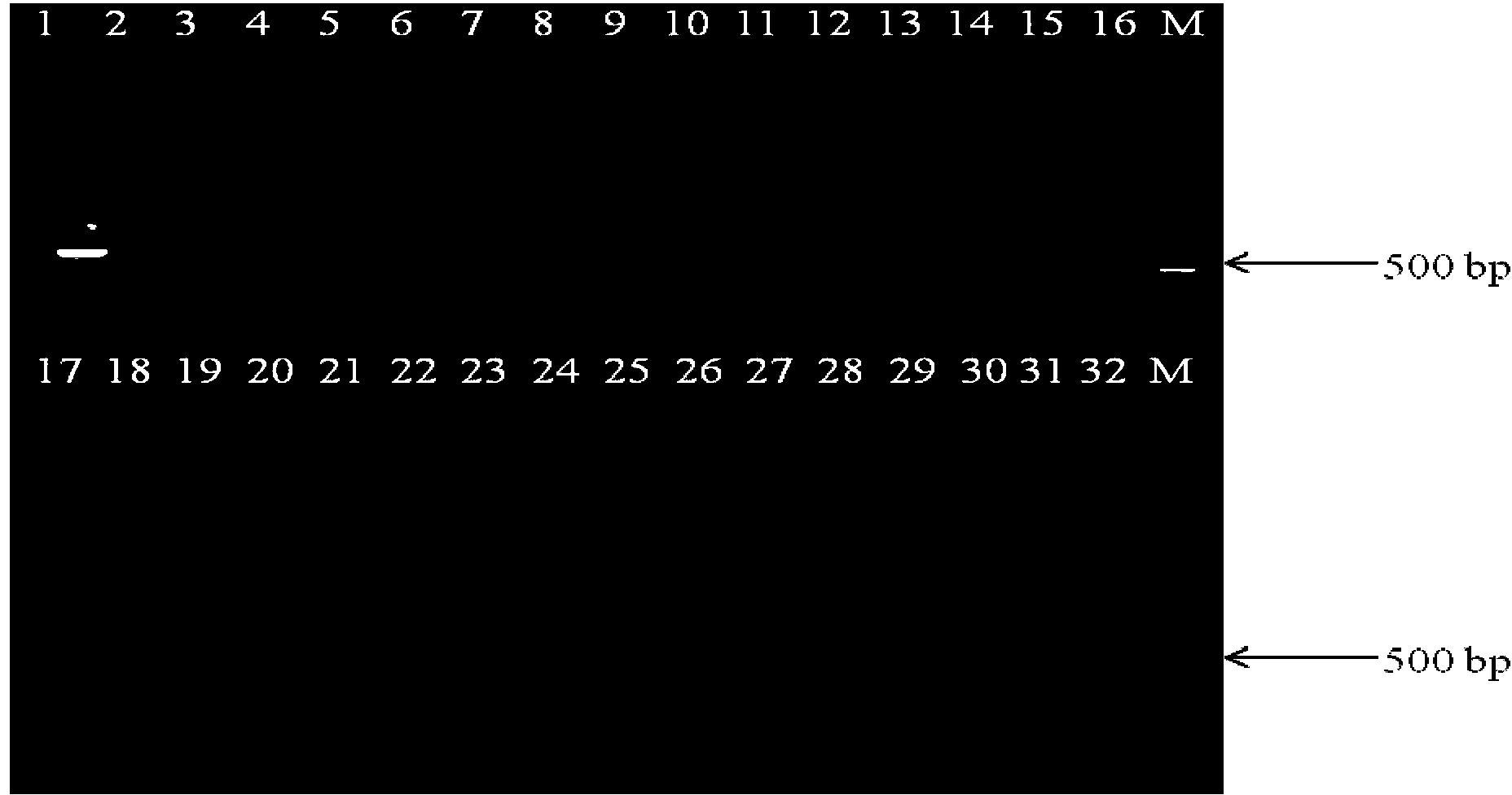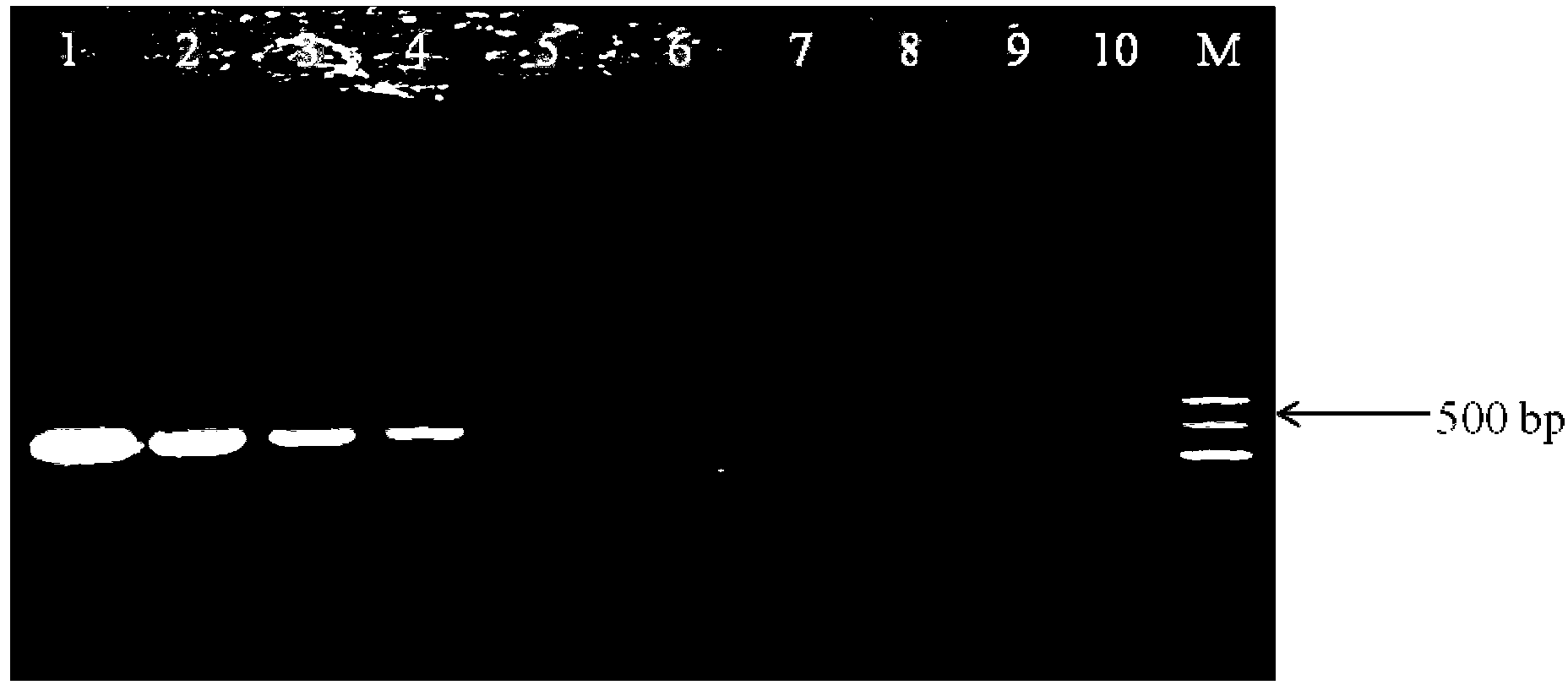Method for detecting cronobacter sakazakii as well as kit and primer thereof
A kit and bacillus technology, applied in the field of microbial detection, can solve the problems of cumbersome operation and time-consuming detection methods, and achieve the effects of high detection efficiency, reliable detection results, and simple result judgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Detection of Cronobacter sakazakii strains
[0028] (1) The standard strain ATCC29544 of the genus Cronobacter sakazakii was detected by PCR using the primer pair and detection method of the present invention.
[0029] The sequences of the primer pairs used are as follows:
[0030] SEN2-L: 5'-ACTGGCTTGGGGCTAATA-3' (SEQ ID NO: 1),
[0031] SEN2-R: 5'-AGAGGCGGATAAATCTTGT-3' (SEQ ID NO: 2).
[0032] Using the above primer pairs, using the genome DNA of the standard strain of Cronobacter sakazakii ATCC29544 as a template, the PCR reaction system and reaction program were established and optimized, and through single factor, multifactor test and hybridization test, a suitable for Detection of the reaction system and reaction parameters of Cronobacter sakazakii strain.
[0033] The PCR reaction system is: 1×PCR reaction buffer, 10-15mmol / L Mg 2+ , 0.2-0.3mmol / L dNTP, 0.1-0.3μM primer SEN2-L, 0.1-0.3μM primer SEN2-R, Taq enzyme 0.05-0.1U / μL, DNA template 10-100ng...
Embodiment 2
[0046] Detection of Artificial Contamination of Cronobacter sakazakii Standard Strain ATCC29544
[0047] Preparation of artificially contaminated samples: After sterilizing 100mL of 10% skim milk solution, take dilutions of 10 -7 , 10 -8 and 10 -9Add 1mL each of the pure culture solution of Cronobacter sakazakii ATCC29544 (440cfu, 44cfu and 4.4cfu) to the above-mentioned sterilized skim milk solution. Place on a shaker, culture at 37°C, 180r / min, sample every 2h, and co-cultivate for 10h. Boiling method (Chen, W., S. Yu, C. Zhang, J. Zhang, C. Shi, Y. Hu, B. Suo, H. Cao, and X. Shi, 2011, Development of a single base extension- tag microarray for the detection of pathogenic Vibrio species in seafood. Applied microbiology and biotechnology 89 (6): 1979-1990.) Genomic DNA was extracted, and PCR reaction was carried out using the aforementioned optimal reaction system and optimal amplification program. The result is as Figure 4 shown by Figure 4 It can be clearly seen tha...
Embodiment 3
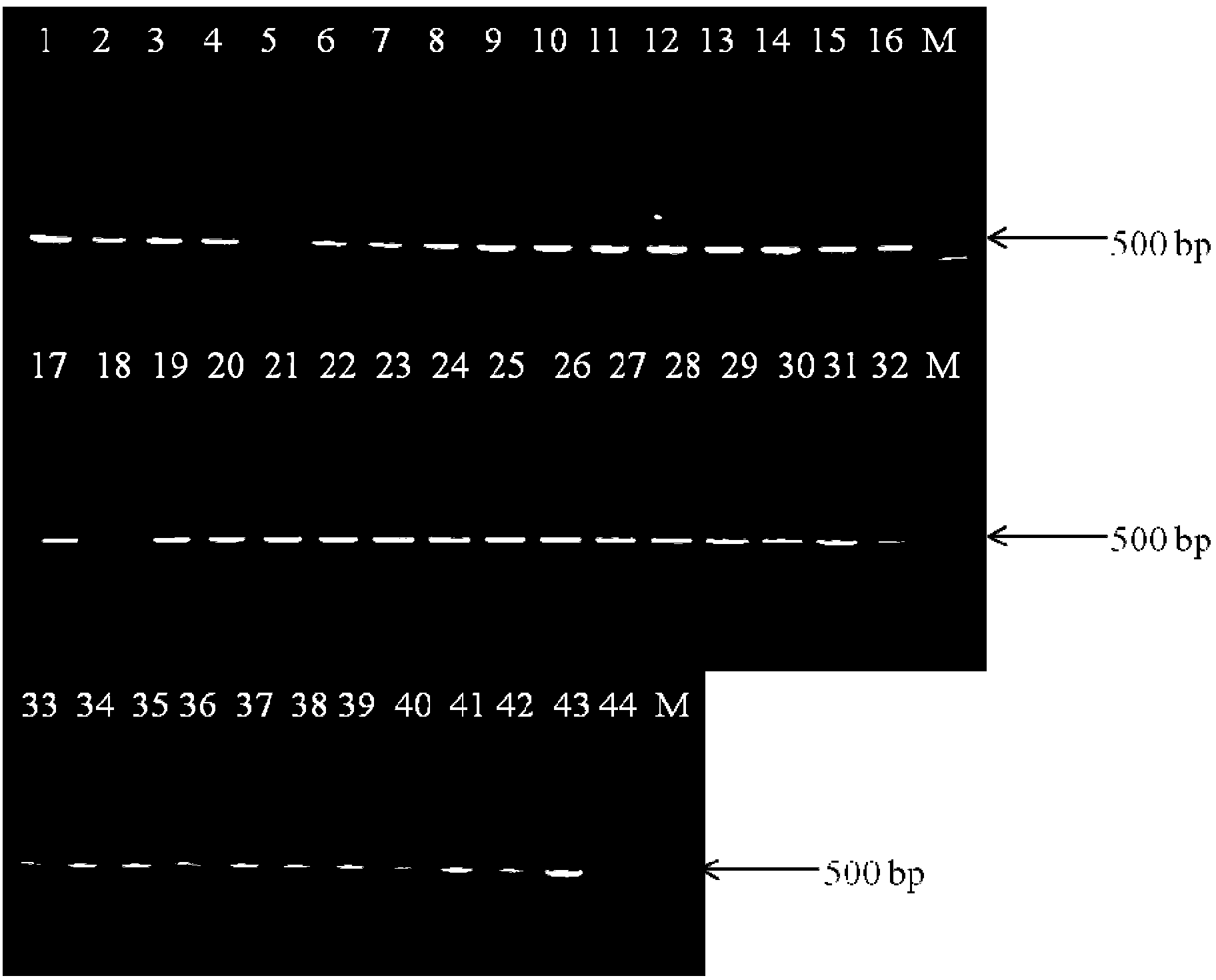
[0049] The 30 food samples (including powdered milk and vegetables) were purchased from farmers' markets and large supermarkets in Shanghai. Add 50g sample to 450mL buffer peptone water (buffer peptone water, BPW, pH8.0) enrichment medium for enrichment culture, after enrichment at 37°C for 18h, take 1mL of each sample into a 1.5mL centrifuge tube, and use Boiling method (Chen, W., S. Yu, C. Zhang, J. Zhang, C. Shi, Y. Hu, B. Suo, H. Cao, and X. Shi, 2011, Development of a single base extension- tag microarray for the detection of pathogenic Vibrio species in seafood. Applied microbiology and biotechnology 89 (6): 1979-1990.) Genomic DNA was extracted, centrifuged at 12,000 rpm / min for 5 min, and 2.5 μL of the supernatant was taken as a PCR template, and sterile Water was used as a negative control, and the aforementioned optimal reaction system and optimal amplification program were used for PCR detection. Each experiment was repeated 3 times. At the same time, 18-24 hours ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com