Total DNA (Deoxyribonucleic Acid) extraction method of bottom mud containing high content of humus at river mouth
A humus and sediment technology, applied in the field of environmental microbiology and molecular biology, can solve the problem of low quality of total DNA in estuary sediment with high humus content, and achieve the effect of easy molecular biology experiment, quality improvement and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] The method of the present invention is used to extract the total DNA of Changjiang Estuary sediment. The sediment of Changjiang Estuary comes from the west side of 200m upstream of the confluence of Changjiang River and Le'an River. The bottom sediment of 0-3 cm in the upper layer is taken as a sample. The sediment sample is gray-black and has a high content of humus. The following are the specific implementation steps:
[0021] 1. Pretreatment of sediment samples: Weigh 10g of estuary sediment into a 50ml centrifuge tube, add 30ml of normal saline, vortex for 5min, let stand for 2min, and centrifuge at 2500r / min for 3min at low speed at 4°C, then 10000r Centrifuge at high speed / min for 8 min, discard the supernatant, and transfer the upper 1 / 2 of the pellet to another new centrifuge tube; repeat the previous step 2 times, transfer the 1.25g sediment to a 10ml centrifuge tube; wash with normal saline The sediment was centrifuged at 10,000 r / min for 8 min at 4°C, and wa...
Embodiment 2
[0029] The method of the present invention is used to extract the total DNA of the Ganjiang estuary sediment. The estuary sediment of the Ganjiang River is from the north of the 150m upstream of the confluence of the Ganjiang River and the Xiu River. The upper layer of 0-3cm sediment is taken as a sample. The sediment sample is yellow clay and contains humus, especially with high pigment content.
[0030]The specific implementation steps are the same as in Example 1.
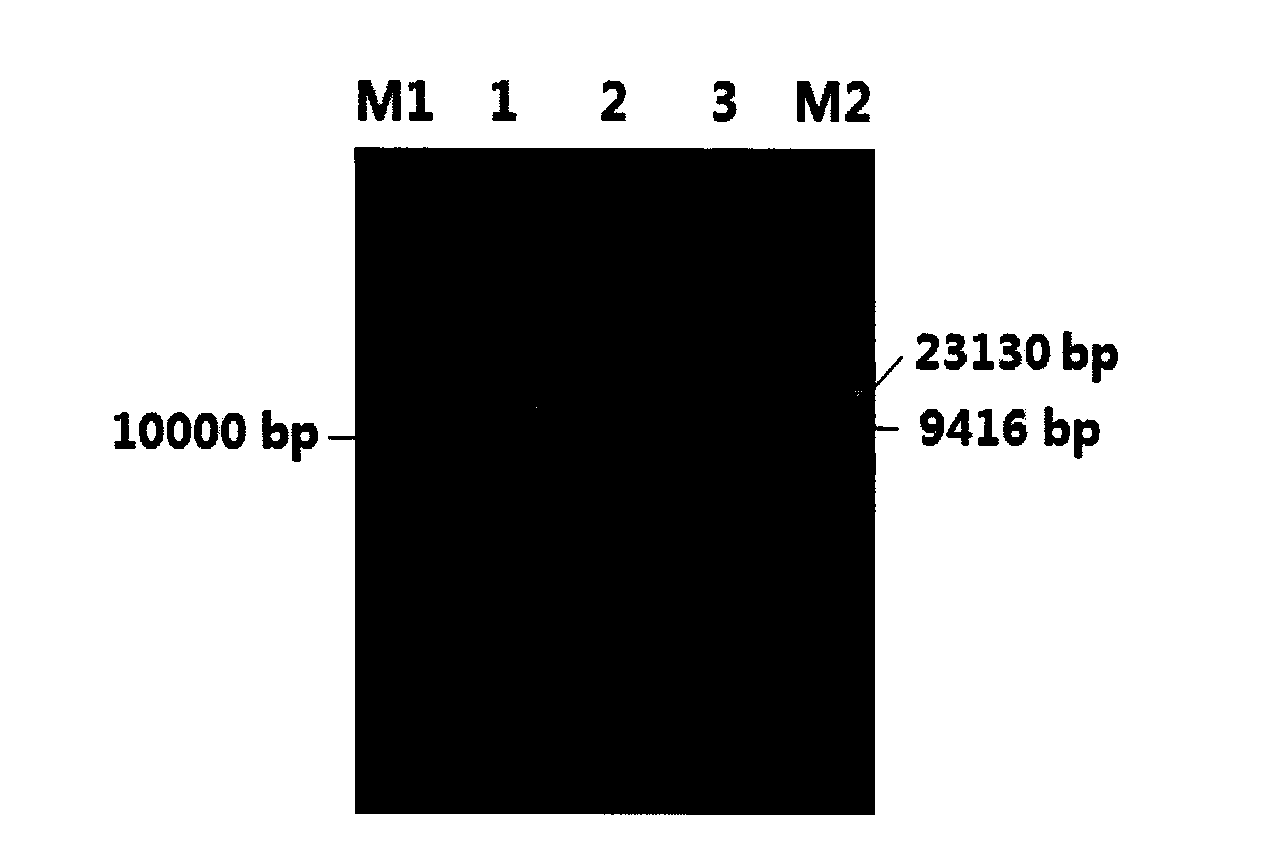
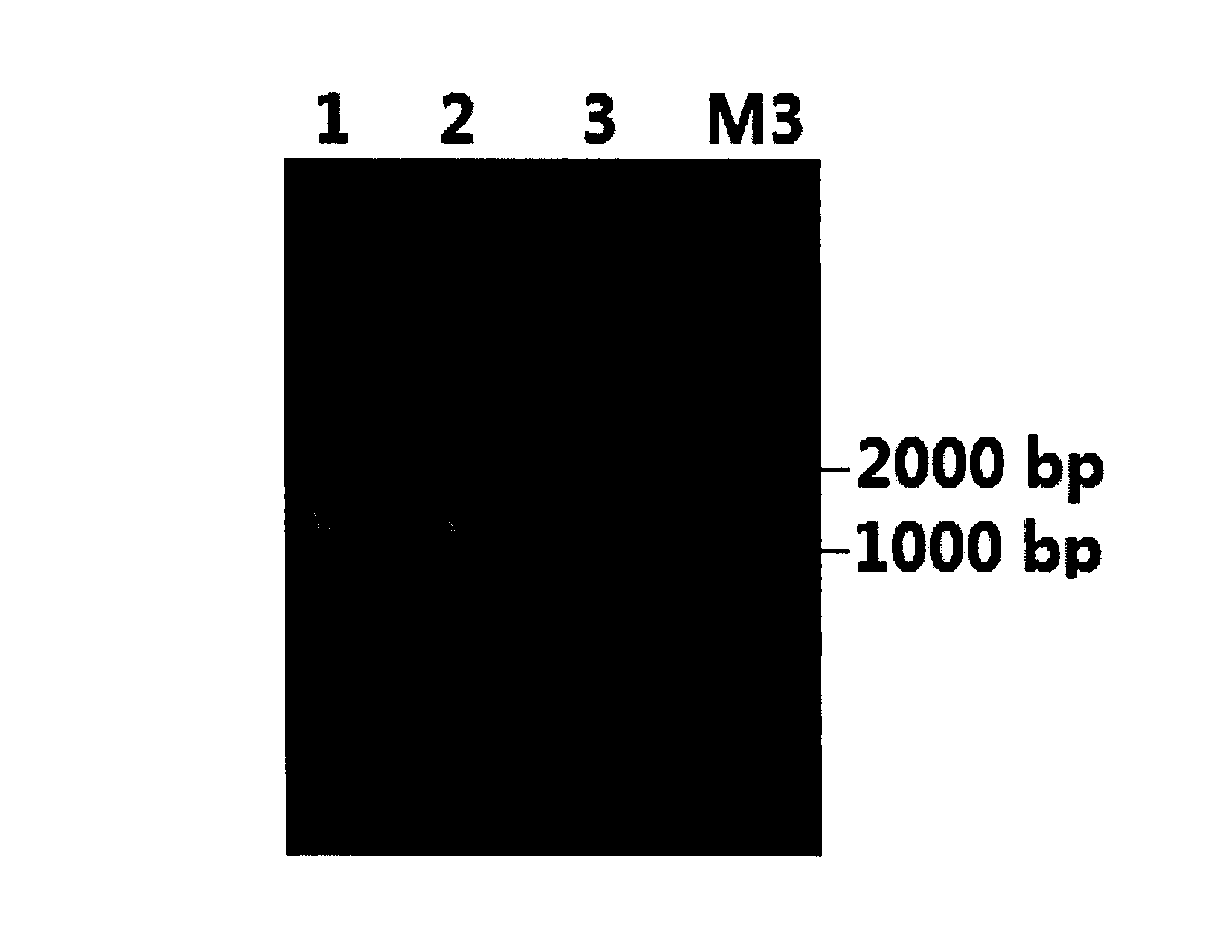
[0031] like figure 1 As shown, the DNA electrophoresis pattern in lane 2 has a single and clear band without tailing, indicating that the DNA is of high purity and has no breaks; such as figure 2 As shown, the electrophoresis map of the PCR product in lane 2 can clearly see the target fragment of about 1380bp, indicating that the method of the present invention is used to extract the total DNA quality of the Ganjiang estuary sediment, which can be used for subsequent molecular biology research.
Embodiment 3
[0033] The soil total DNA is extracted by the method of the present invention. The soil was from Meiling, Nanchang City, Jiangxi Province. The topsoil on the mountain with a lot of fallen leaves and decay was taken as a sample. The soil sample is gray-black and contains a lot of humus.
[0034] The specific implementation steps are the same as in Example 1.
[0035] like figure 1 As shown in Figure 3, the DNA electrophoresis pattern in lane 3 has a single and clear band, and no tailing phenomenon, indicating that the DNA is of high purity and no breakage; such as figure 2 As shown in the electrophoresis map of the PCR product in lane 3, the target fragment of about 1380bp can be clearly seen, indicating that the total DNA extracted from Meiling soil by the method of the present invention is of high quality and can be used for subsequent molecular biology research.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com