Method for decolorizing nucleotide enzymatic hydrolysate
A nucleotide enzymatic solution and pigment technology, applied in the field of separation and purification of 5'-nucleotides, can solve problems affecting the purity and chroma of nucleotide products, reduce the reuse rate of resins, resin poisoning, etc., and achieve Improve appearance color, low price, large adsorption effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1: Preparation of nucleotide enzymatic hydrolysis solution.
[0048] The RNA enzymatic hydrolysis process uses nuclease p1 to act on the RNA solution to obtain 4 kinds of 5'-mononucleotides, namely 5'-adenylic acid (AMP), 5'-cytidylic acid (CMP), 5'- Guanylate (GMP) and 5'-Uridylic acid (UMP). Specific steps are as follows:
[0049] (1) Prepare 600ml of 6% RNA solution with pH=5.5: first use 1mol / L NaOH solution to adjust the pH to about 13, then add RNA powder (36g) to dissolve, then use 1mol / L NaOH solution to adjust the pH to 5.5, place in a 70°C water bath and preheat to 70°C.
[0050] (2) Add 50ml of enzyme solution preheated at 70°C for half an hour to the RNA solution, shake well, and enzymatically digest at 70°C for 3h50min.
[0051] (3) Add 0.2% (1.2g) activated carbon and continue enzymatic hydrolysis for 50 minutes.
[0052] (4) Cool to 45° C., centrifuge at 4000 rpm for 10 minutes, and filter with suction to obtain RNA enzymatic hydrolysis sol...
Embodiment 2
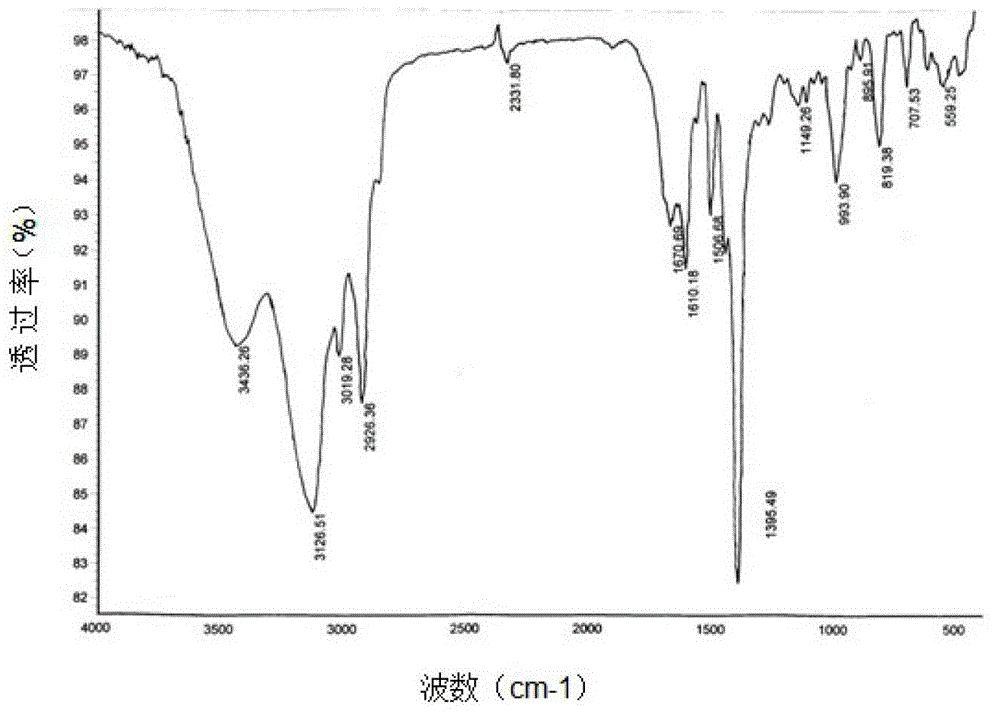
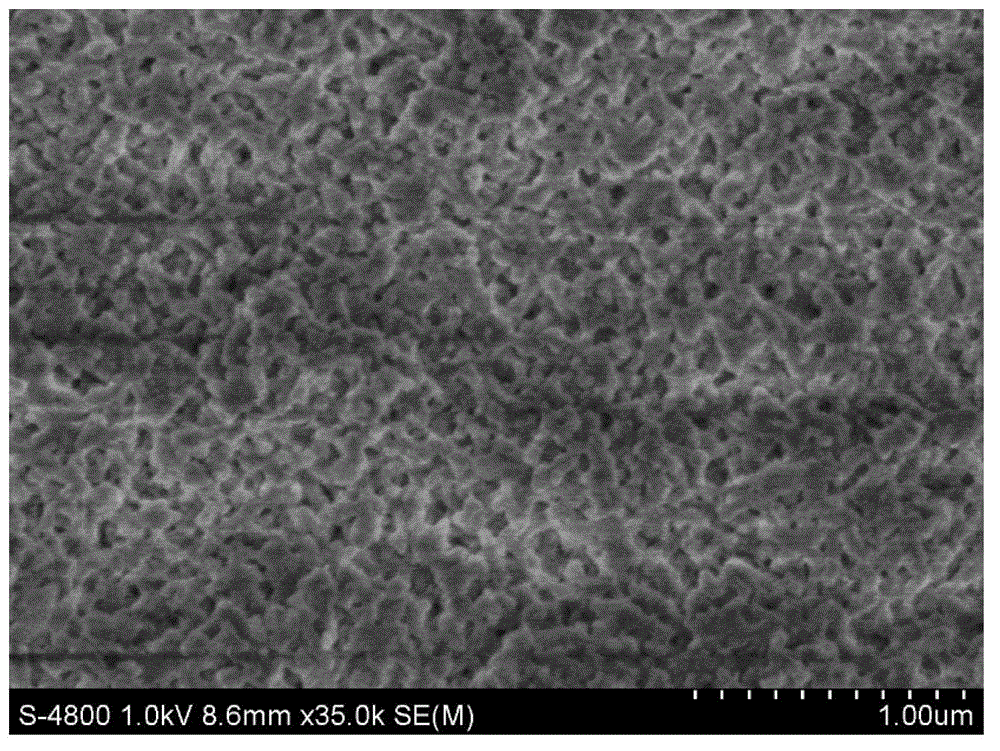
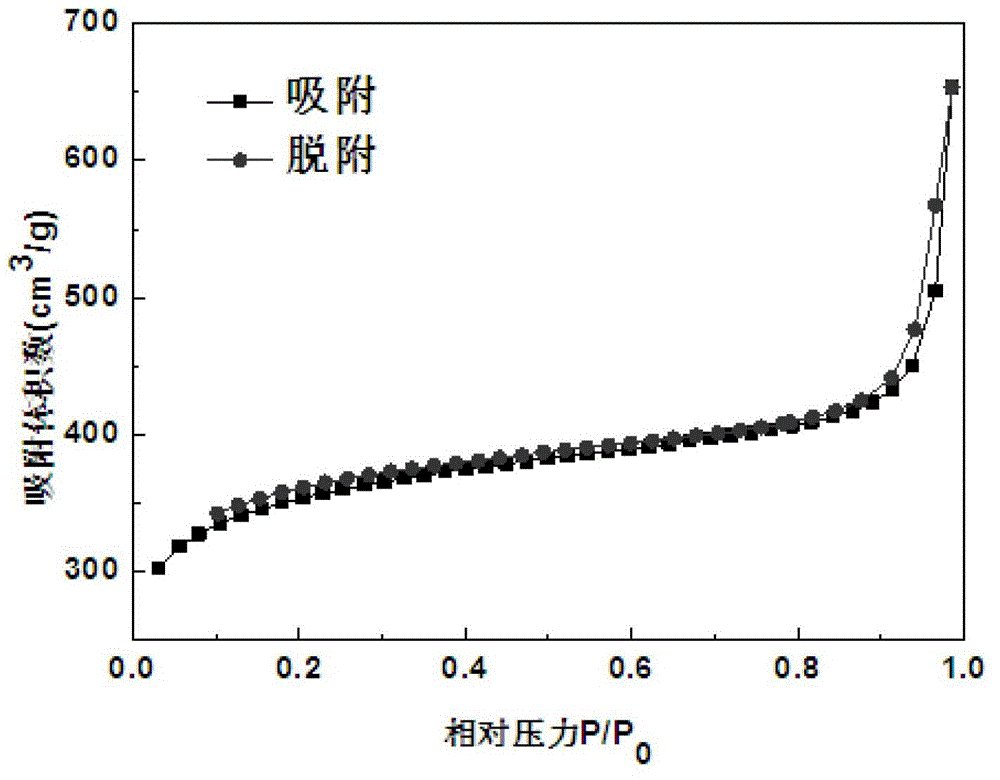
[0053] Embodiment 2: the acquisition of ultra-high cross-linked resin SX-01.
[0054] In a clean and dry 2000mL three-neck flask, add 200g of dry chloromethylated polystyrene resin (chlorine balls), add 1200g of o-nitroethylbenzene, stir and mix well, fully swell at room temperature for more than 2 hours, stir and add Methylamine 4 times the weight of chlorine balls, then add 20g of anhydrous ferric chloride as a catalyst, program temperature rise to 135-150°C under stirring, and then carry out Friedel-Crafts post-crosslinking reaction at this temperature for a certain period of time to control the residual chlorine content At 1-3%, stop the reaction, suck out the mother liquor after cooling, and repeatedly extract the resin with industrial alcohol until the extraction effluent is colorless and clear, then wash the resin with 4% hydrochloric acid and distilled water until it is neutral, and dry it for later use.
[0055] The obtained product is in the shape of brown round part...
Embodiment 3
[0056] Embodiment 3: the pretreatment of ultrahigh cross-linking resin SX-01.
[0057] Rinse the resin with 2BV of ethanol at a flow rate of 2BV / h→wash away the ethanol with deionized water→2BV0.1mol / L HCl solution wash the resin at a flow rate of 1BV / h→wash with water until neutral→use 2BV0.1mol / L Rinse the resin with NaOH solution at a flow rate of 1BV / h→wash with water to neutral→wash the resin with 2BV0.1mol / L HCl solution at a flow rate of 1BV / h→wash with water to neutral.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com