Preparation method and application of colon cancer cell strain expressed by stabilized silent PKM2 gene
A colon cancer cell, gene expression technology, applied in the field of colon cancer cell line preparation, can solve the problem of unclear tumor and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] (1) Design of shRNA oligonucleotide sequence targeting PKM2 gene
[0019] The complete mRNA sequence of the PKM2 gene (NM-001206796) was found in GenBank, and the specificity was confirmed by Blast homology comparison, and the secondary structure of the target mRNA sequence was evaluated by using RNA Structure4.4 software, and finally a 21nt target was screened. Nucleotide sequence: SEQ ID NO: 5, CGGGTGAACTTTGCCATGAAT, the target position is 946-966.
[0020] Design and synthesize a shRNA DNA template single strand according to the target nucleotide sequence, named shRNA1, and design a pair of control sequences shRNAc, the specific sequence is shown in Table 1. The designed shRNA oligonucleotide chains were synthesized by Shanghai Yingjun Biotechnology Co., Ltd.
[0021] Table 1 shRNA oligonucleotide sequences targeting PKM2 gene
[0022]
[0023] (2) Construction of shRNA lentiviral expression vector
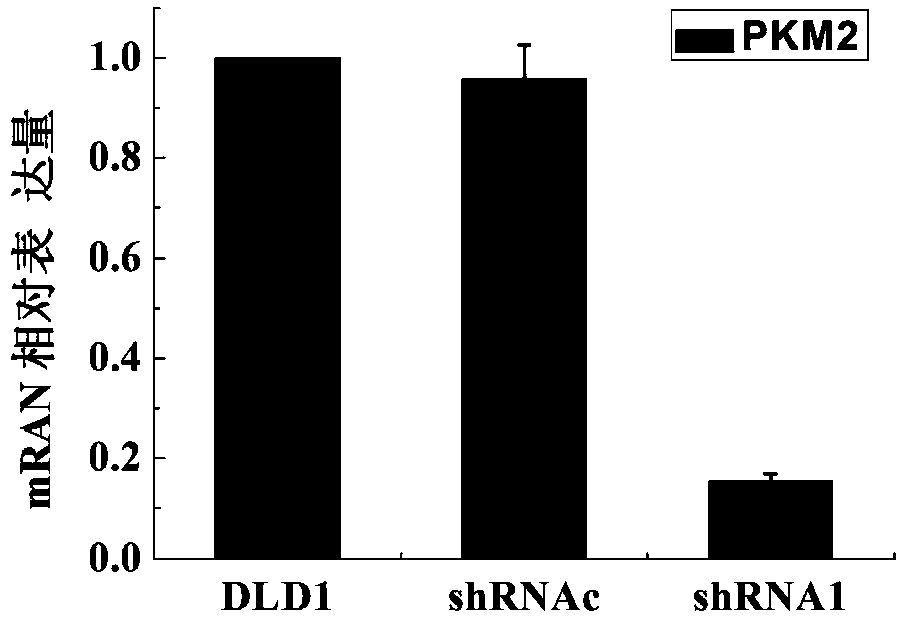
[0024] Mix the synthetic shRNA oligonucleotide single-strande...
Embodiment 2
[0038] Example 2 Fluorescent quantitative PCR detection of glutamine metabolism-related enzymes and receptor gene expression
[0039] Using each cDNA in Example 1 (5) as a template, using GAPDH as an internal reference, fluorescent quantitative PCR was used to detect the relative expression of glutamine metabolism-related enzyme genes, and the reaction conditions were set: 94°C for 30s, one cycle; 94°C for 5s ; 60°C for 34s, 40 cycles. The result is as Figure 4 As shown, compared with shRNAc cells, the gene expression levels of glutamine metabolism-related enzymes and receptors in shRNA1 cells increased, indicating that silencing the expression of PKM2 gene caused the compensatory effect of glutamine in cells.
Embodiment 3
[0040] Example 3 MTT method (thiazolium blue colorimetric method) detection of glutamine compensation in stable cell lines
[0041] Inoculate 8×10 shRNAc cells and shRNA1 cells in good growth state, respectively. 3 Put each well into a 96-well plate, incubate in a 37°C incubator for 24 hours, add Don (glutaminase inhibitor) at a final concentration of 0.5mM, set up five duplicate wells, and use the medium without drug as a control . After 48 hours of drug action, add 20 μL of 5 mg / mL MTT solution to each well, continue to incubate for 4 hours, and terminate the culture. Carefully aspirate and discard the culture supernatant in the wells, add 150 μL DMSO to each well, and shake for 10 minutes to fully dissolve the purple crystals. At 570nm wavelength on the enzyme-linked immunoassay instrument, measure the light absorption value (A value) of each hole, the result is as follows Figure 5 As shown, the survival rate of shRNA1 cells is lower than that of control cells, which sh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com