Diamondback moth peptidoglycan recognition protein, preparation method and application thereof
A technology for moth peptidoglycan and recognition protein, which is applied in the field of diamondback moth peptidoglycan recognition protein and its preparation, and can solve the problems of no related reports and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
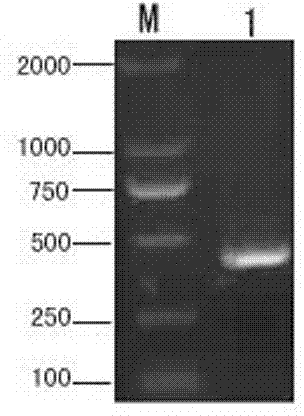
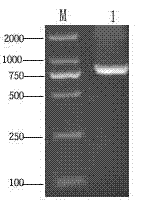
[0028] Example 1 Cloning of Plutella xylostella peptidoglycan recognition protein (PGRP-S1) gene
[0029] S1. laboratory rearing diamondback moth ( Plutella xylostella ) larvae, Paecilomyces fumigatus ( Paecilomyces Fumosoroseus ) as the tested strain.
[0030] S2. Determination of the transcriptome sequence of Plutella xylostella: Collect the 1st to 4th instar larvae, prepupa, pupae, and adults of Plutella xylostella, extract the total RNA, and use RNA-seq technology to determine the transcriptome sequence. The extraction method of total RNA refers to Trizol (Invitrogen, USA) kit manual operation. A total of 34,522,812 clean reads were detected, the average length of the reads was 90bp, and 3,107,053,080 bases were obtained. Finally, 107710 Unigenes were assembled, with a length ranging from 200 to 3000 bp. After COG, GO, KEGG annotation and analysis, a total of 68,984 Unigenes with high annotation reliability were obtained, and 725 Unigenes directly related to inse...
Embodiment 2
[0050] Example 2 Temporal and spatial expression pattern of Plutella xylostella peptidoglycan recognition protein PGRP-S1
[0051] S1. Collect Plutella xylostella at different development stages, including eggs, 1st instar larvae; 2nd instar larvae; 3rd instar larvae; 4th instar larvae;
[0052] S2. The 4th instar larva was dissected, and the immune organs of fat body, blood cells, epidermis, midgut and Malpighian duct were collected respectively.
[0053] S3. Collect Plutella xylostella at different ages and tissues, add liquid nitrogen, use TAKARA’s RNAiso Plus extraction reagent, and follow the instructions to obtain total RNA at different ages and tissues.
[0054] S4. Synthesize cDNA of different ages according to the RACE kit instructions of CLONTECH company, add the following reagents at a time: 10 micrograms of total RNA, add 20 microliters of reaction solution and react at 42°C for 90 minutes. The reaction was terminated at 72°C for 10 minutes. Reaction composition:...
Embodiment 3
[0062] Example 3 Plutella xylostella PGRP-S1 is against Paecilomyces fumigatus ( P. fumosoroseus ) of the recognition effect
[0063] S1. Get the 4th instar Plutella xylostella larvae, collect the spores of I. fumigatus and dilute to 10 with sterile water. 5 Spores / mL, inject diamondback moth with a micro-syringe, about 8uL per head, and take 6 diamondback moths at 0h, 3h, 6h, 12h, 18h, 24h and 36h, respectively. Total RNA was extracted, cDNA was reverse transcribed, and it was used as a template to detect the expression of PGRP-S1 at different time points after infection by I. fumigatus. The experiment was repeated 3 times.
[0064] S2. Using the β-Actin gene (AB282645) of Plutella xylostella as an internal reference gene, fluorescent quantitative PCR primers were designed according to the sequences of PGRP-S1 and β-Actin of Plutella xylostella. The quantitative primer sequence for PGRP-S1 is:
[0065] qF: 5'-TCATCAACCACTCCGTGTCCCCC-3' (shown in SEQ ID NO: 17).
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com