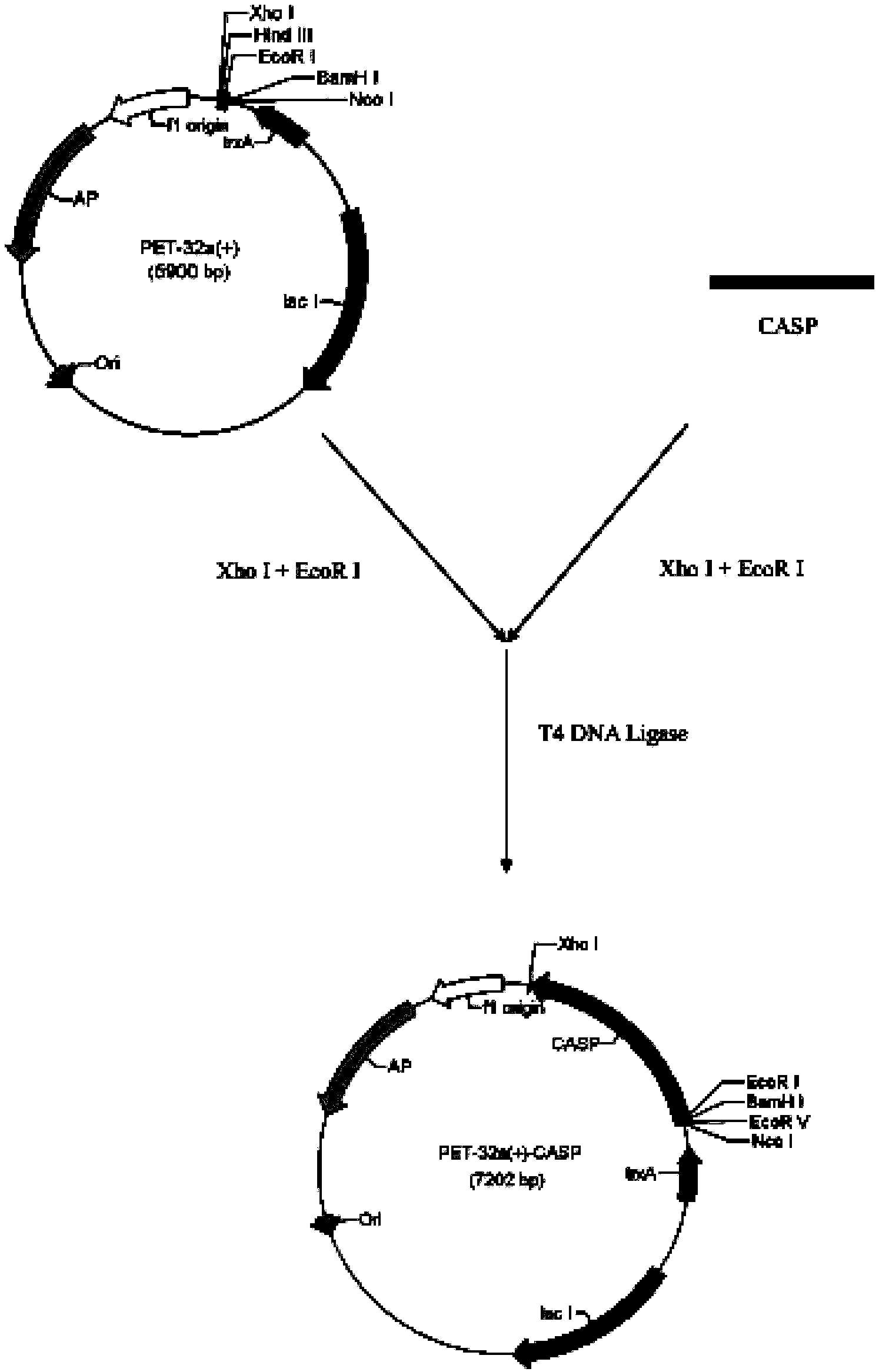
Branchiostoma belcheri chitin-binding associated serine protease CASP gene for identifying chitin and application thereof
A technology of serine protease and amphioxus, applied in CASP protein and drug application field, can solve problems such as complement pathway damage, lack of hemolytic activity, suppurative infection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Extraction of amphioxus total RNA and amplification of CASP partial fragments.
[0053] Extraction of total RNA and synthesis of RACE cDNA: Whole amphioxus was taken, total RNA was extracted with Trizol reagent, and protein was extracted by phenol / chloroform extraction to obtain total RNA of amphioxus. Take 1 μg of total RNA, and perform reverse transcription to synthesize the first strand according to the instructions of TOYOBO's First Strand cDNA Synthesis Kit ReverTra Ace-α-TM (code No. FSK-100). Take 2 μl of the first-strand cDNA product, and use the combined primers 5′PCR Primer: 5′-ACCCATCCCTCCCAGTCAC-3′ and 3′PCR Primer: 5′-GTAGACACTCGGCTTGGCG-3′ designed according to the predicted sequence of the CASP gene in the Florida amphioxus sequencing database as primers, RT-PCR was carried out to amplify the 899bp partial fragment of CASP. The amplified target fragment was connected to pGEX T easy vector (promega) and transformed into DH5α Escherichia coli, a...
Embodiment 2
[0054] Example 2: RACE amplifies the 5' and 3' ends of the CASP gene.
[0055] According to Invitrogen's GeneRacerTM Kit, perform RNA dephosphorylation RACE, decapping reaction, RNA oligo ligation and mRNA reverse transcription to synthesize a cDNA strand, and use Takara's LA Taq polymerase to perform PCR amplification according to the reaction system of GeneRacerTM Kit . Among them, according to the sequence of the CASP partial fragment amplified above, the gene-specific outer primer for 5′ RACE amplification is designed as 5′-gene-specific primer: 5′-GACCATCCAAGGCATCACCAGTT-3′; the inner nest primer is 5′ -nested primer: 5′-GGACACTGACATGGACTGAAGGAGTA-3′; CASP’s 3′ RACE amplified gene-specific coat primer is 5′-gene-specific primer: 5′-GGAAGGAGACCACCCACGGCT-3′; inner nest primer is 5′-nested primer: 5'-CGCTACGTAACGGCATGACAGTG-3'. The amplified target fragment was connected to pGEX T easy vector (promega), transformed into DH5α Escherichia coli, and the recombinant clone was...
Embodiment 3
[0056] Example 3: Amplification and sequence analysis of the full length of CASP.
[0057] According to the sequence of the CASP gene spliced in Example 2, primers 5'-TCTGCATTTCATCATCAAACG-3' and 5'-TTGGGTGAGGTTTCTAAGGCTAA-3' were designed, and the whole gene of CASP was amplified by Takara's LATaq polymerase, and the obtained 1976 fragment was amplified. Electropherogram such as figure 2 shown. The amplified target fragment was connected to pGEX T easy vector (promega) and then transformed into DH5α Escherichia coli, and the recombinant clone was selected for sequencing. Blast homology analysis showed that the EST sequence encoding the whole gene of CASP was obtained.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com