Preparation method and application of penicillium genetically engineering bacterium
A technology of genetically engineered bacteria and Penicillium, applied in the field of genetic engineering, can solve problems such as non-expression of genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: Construction of overexpression vector
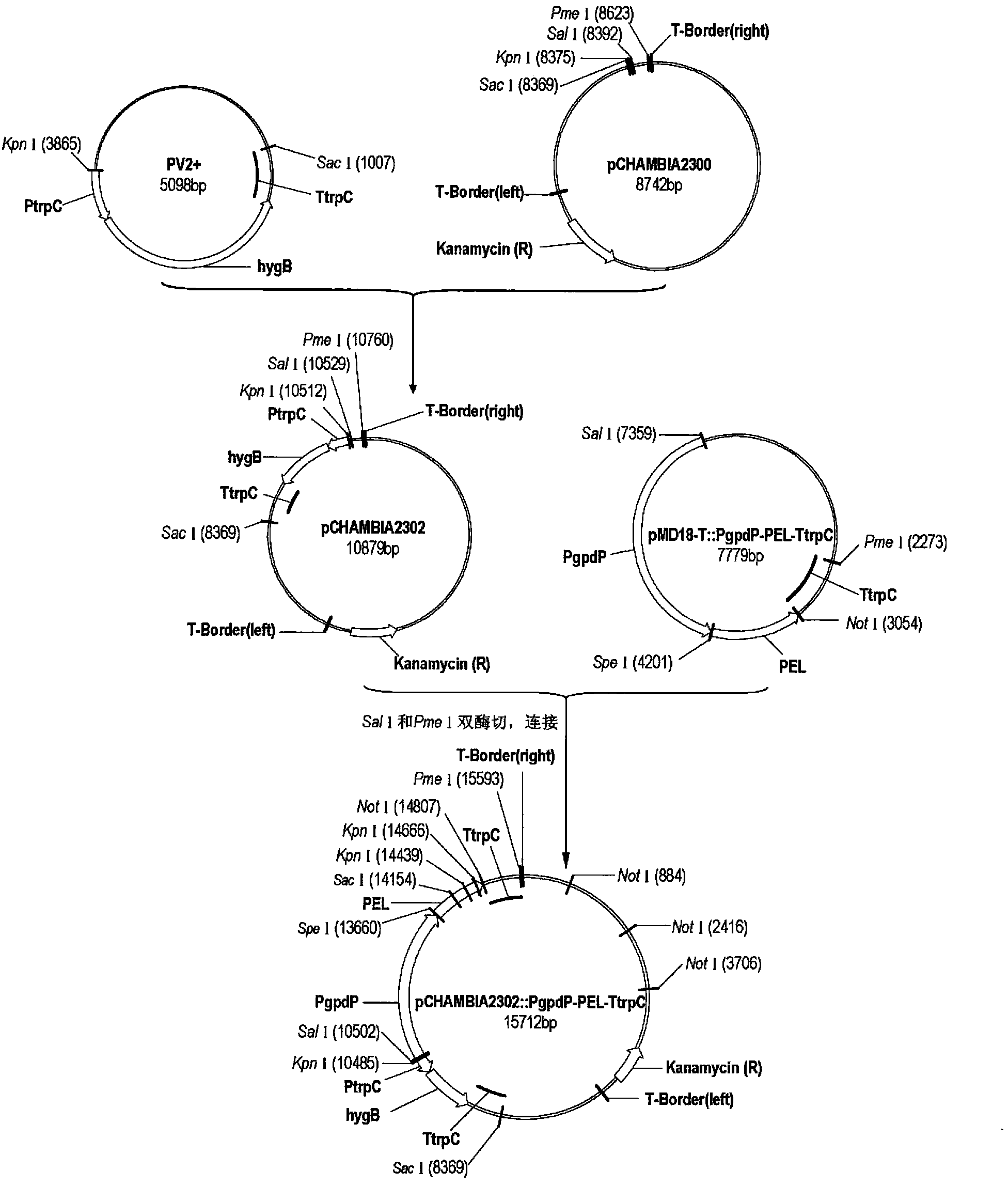
[0065] See figure 1 , the detailed steps are as follows:
[0066] (1) utilize restriction endonuclease Sac I, Kpn I to hygromycin resistance expression cassette from plasmid PV2 + The fragments were excised by the upper enzyme, and the fragments were recovered by gel, and cloned into the corresponding restriction site of the pCAMBIA2300 plasmid to obtain the recombinant plasmid pCHAMBIA2302 with hygromycin resistance; the hygromycin expression cassette can also be derived from any other DNA containing this sequence or synthesized directly. In addition to pCAMBIA2300, plasmids used to construct PEL gene overexpression vectors can also be used to express plasmids in filamentous fungi, such as pCAMBIA1300, pCAMBIA3300 and other pCAMBIA series vectors and pHB vectors.
[0067] (2) Take 0.2 g of Penicillium extensa mycelium and grind it with liquid nitrogen, add 1 mL of high-salt and low-pH extract, bathe in 65 ° C for...
Embodiment 2
[0074] Embodiment 2: the acquisition of Penicillium genetically engineered bacteria
[0075] The detailed steps are as follows:
[0076] (1) Pick isolated and purified wild-type Penicillium and inoculate it on a PDA plate, culture it at 28° C. for about 20 days, and wash the mature spores with sterile water.
[0077] (2) Inoculate the engineering Agrobacterium EHA105 containing the final vector pCHAMBIA2302::PgpdP-PEL-TtrpC in Example 1 in the LB liquid medium containing 100 μg / mL streptomycin and 100 μg / mL kanamycin at 28° C., 200 rpm Cultivate overnight, reactivate with MM medium containing 100 μg / mL streptomycin and 100 μg / mL kanamycin, and culture at 28°C, 220 rpm for 48 hours. Draw an appropriate amount of culture and centrifuge at 5000rpm to remove the supernatant, wash with IM medium, and finally dilute to OD with IM medium 600 =0.15, then cultured at 28°C and 220rpm for 6-8 hours until OD 600 = 0.5 to 0.6.
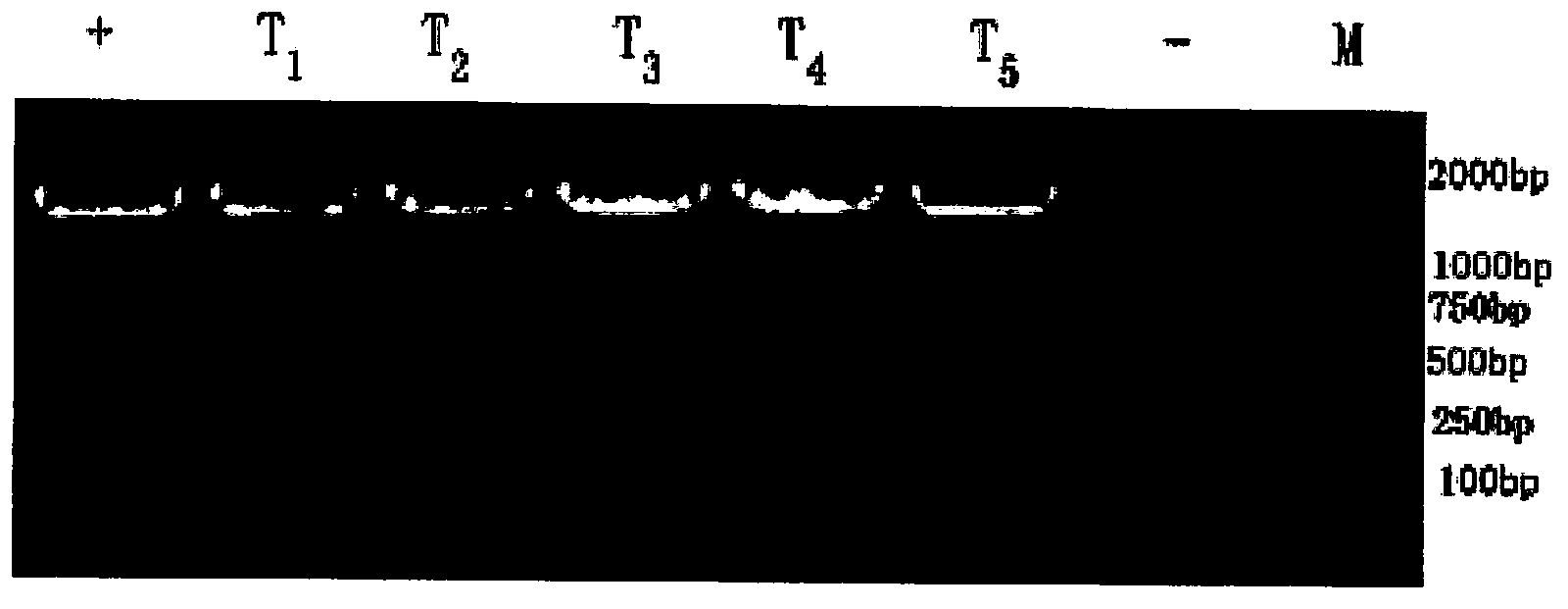
[0078] (3) The fresh spores obtained in step (1) are prep...
Embodiment 3
[0084] Embodiment 3: producing lipase ability comparison
[0085] The experiment randomly selected the Penicillium genetically engineered bacterium obtained in Example 2 to inoculate on the PDA medium, and after cultivating for 10 days, insert them into 50 mL of the seed medium, cultivate them on a shaker at 28° C. for 24 hours at 210 rpm, and then transfer them to 10% of the inoculum. Put into the fermentation medium (250mL Erlenmeyer flask with 30mL fermentation medium), ferment at 28°C and 210rpm for 48h; then centrifuge the fermentation broth, and take the supernatant to detect lipase activity.
[0086] Use the acid-base titration method to detect the lipase activity. The detection steps are: take 20 100mL Erlenmeyer flasks, add 4.0mL Gly-NaOH buffer solution of pH 9.4 and 5.0mL olive oil emulsion; Preheat the water bath at 36°C for 5 minutes. After filtering the enzyme solution, dilute it with 0.05mol / L Gly-NaOH buffer solution with pH9.4 to make the enzyme activity 4~5u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com