Application of abca3 gene in the preparation of congenital ccmc diagnostic products
A technology for congenital cataracts and products, applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of limited understanding of disease-causing genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: Screen the mutation site of ABCA3 gene from congenital CCMC family
[0065] 1. Extraction of peripheral blood genomic DNA:
[0066] In compliance with the relevant national policies and regulations, and on the basis of the consent of the sampling subject, draw 2-5ml of peripheral venous blood from the first family member, put it into an EDTA anticoagulant tube, and store it at -80°C for later use; the frozen EDTA anticoagulant blood After melting at room temperature, put 500 μL into a centrifuge tube, add an equal volume of TE (pH 8.0), mix well, centrifuge at 10,000 rpm for 10 minutes at 4°C, and discard the supernatant.
[0067] Add 180 μLTE, 20 μL SDS (10%), 8 μL proteinase K (10 mg / ml), mix well, and place in a 37°C water bath overnight. Remove the sample from the water bath and briefly centrifuge to pellet the sample. Add an equal volume of Tris-saturated phenol (about 300 μL) to the reaction tube, mix thoroughly, centrifuge at 10,000 rpm for 10 min...
Embodiment 2
[0122] Example 2: Screening the mutation site of ABCA3 gene from another congenital CCMC family
[0123] 1. Extraction of genomic DNA from peripheral blood: the conventional phenol-chloroform method described in Example 1 was used to extract genomic DNA from the peripheral blood of each member of the family.
[0124] 2. PCR amplification target fragment: reaction conditions and reaction system:
[0125] (1) PCR reaction conditions: 94°C for 3min; 94°C for 40sec, 55±3°C for 40se, 72°C for 60sec, 30-35cycle; 72°C for 10min.
[0126] (2) Reaction system: (TAKARALATaqpolymerase)
[0127]
[0128] The reaction system was used to amplify each patient's genomic DNA template and 19 pairs of primers for the ABCA3 gene.
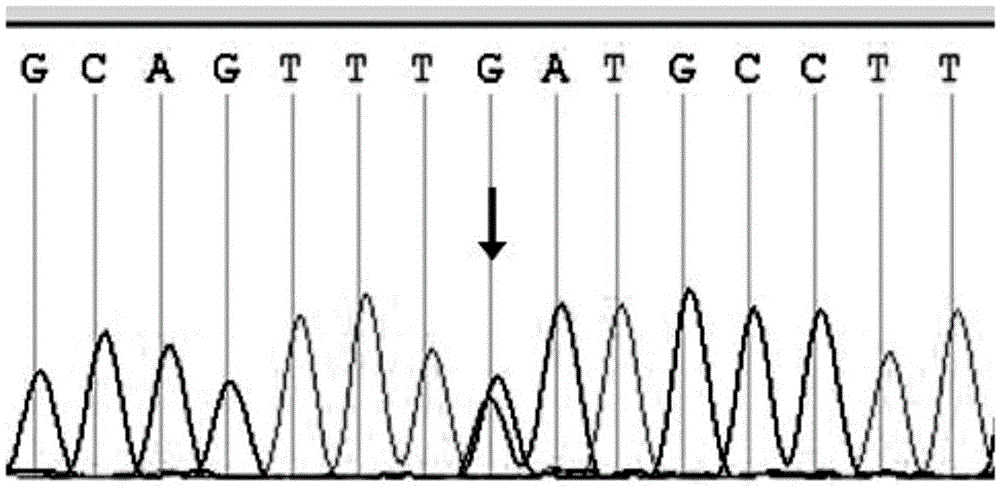
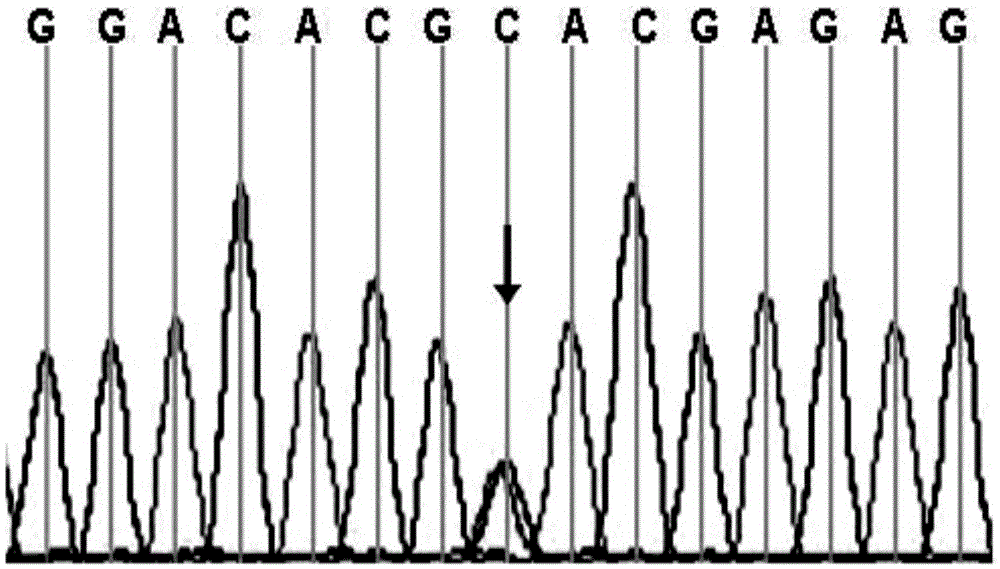
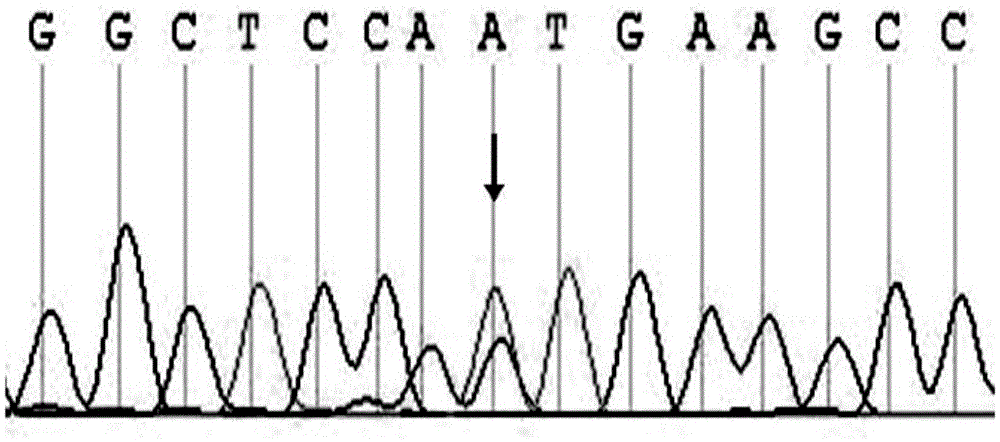
[0129] 3. Sequencing of PCR products: The genomic DNA templates of the patients in the family and the amplification products of 19 pairs of primers of the ABCA3 gene were sequenced using the conventional Sanger sequencing method. A heterozygous mutation occurred ...
Embodiment 3
[0130] Example 3: Screening the mutation site of ABCA3 gene from another 2 patients with congenital sporadic CCMC
[0131] Collection of sporadic patients with CCMC: A total of 2 sporadic patients with definite diagnosis of CCMC were collected in Shandong Eye Institute and Qingdao Eye Hospital.
[0132] Genomic DNA extraction from peripheral blood:
[0133] Take 2-5ml of peripheral venous blood from 2 patients with sporadic CCMC, put them into EDTA anticoagulant tubes, and freeze them at -80°C for later use; after thawing the frozen EDTA anticoagulated blood at room temperature, put 500 μL into centrifuge tubes, and use them for implementation. The conventional phenol-chloroform method described in Example 1 was used to extract genomic DNA from the peripheral blood of each member of the family.
[0134] PCR amplification target fragment: reaction conditions and reaction system:
[0135] (1) PCR reaction conditions: 94°C for 3min; 94°C for 40sec, 55±3°C for 40se, 72°C for 60s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com