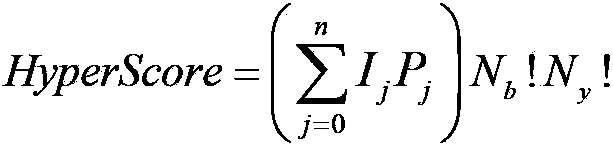Method for identification of protein by utilizing high-energy collision induced ionization dissociation technology
A protein and high-energy technology, applied in the field of bioinformatics, can solve problems such as limited modification types, no pre-screening method for HCD data modification types, optimization of the construction method of the library and the search engine of the library, etc., to improve the identification success rate, The effect of improving identification sensitivity and improving peptide identification sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Using HCD tandem MS / MS data to analyze and identify peptide sequences
[0037] 1. Enzymatic hydrolysis to obtain peptides
[0038] Saccharomyces cerevisiae strain ATCC 201388 (BY4741, MATa his3delta1 leu2delta0 met15delta0ura3delta) was purchased from American Type Culture Collection.
[0039] Strain culture: use YPD medium to cultivate yeast ATCC 201388, culture on a constant temperature shaker at 30°C until OD600 is 1.5, collect the bacteria by centrifugation at 5000rpm for 5 minutes, pour out the supernatant, and rinse the precipitate with 0.1% sodium azide phosphate buffer After centrifuging to remove the supernatant, the cells were collected and stored in a -80°C refrigerator.
[0040] Cell lysis: add urea lysate (8M urea, 50mM ammonium bicarbonate, 50mM iodoacetamide) to the yeast cell precipitation, then add glass beads equal to the cell volume, and place it on a vortex mixer at the maximum speed Vortex to lyse for 5 minutes, centrifuge at 13000 rpm ...
Embodiment 2
[0068] Example 2. Optimization of Peptide Sequence Analysis and Identification Using HCD Tandem MS / MS Spectrogram Data
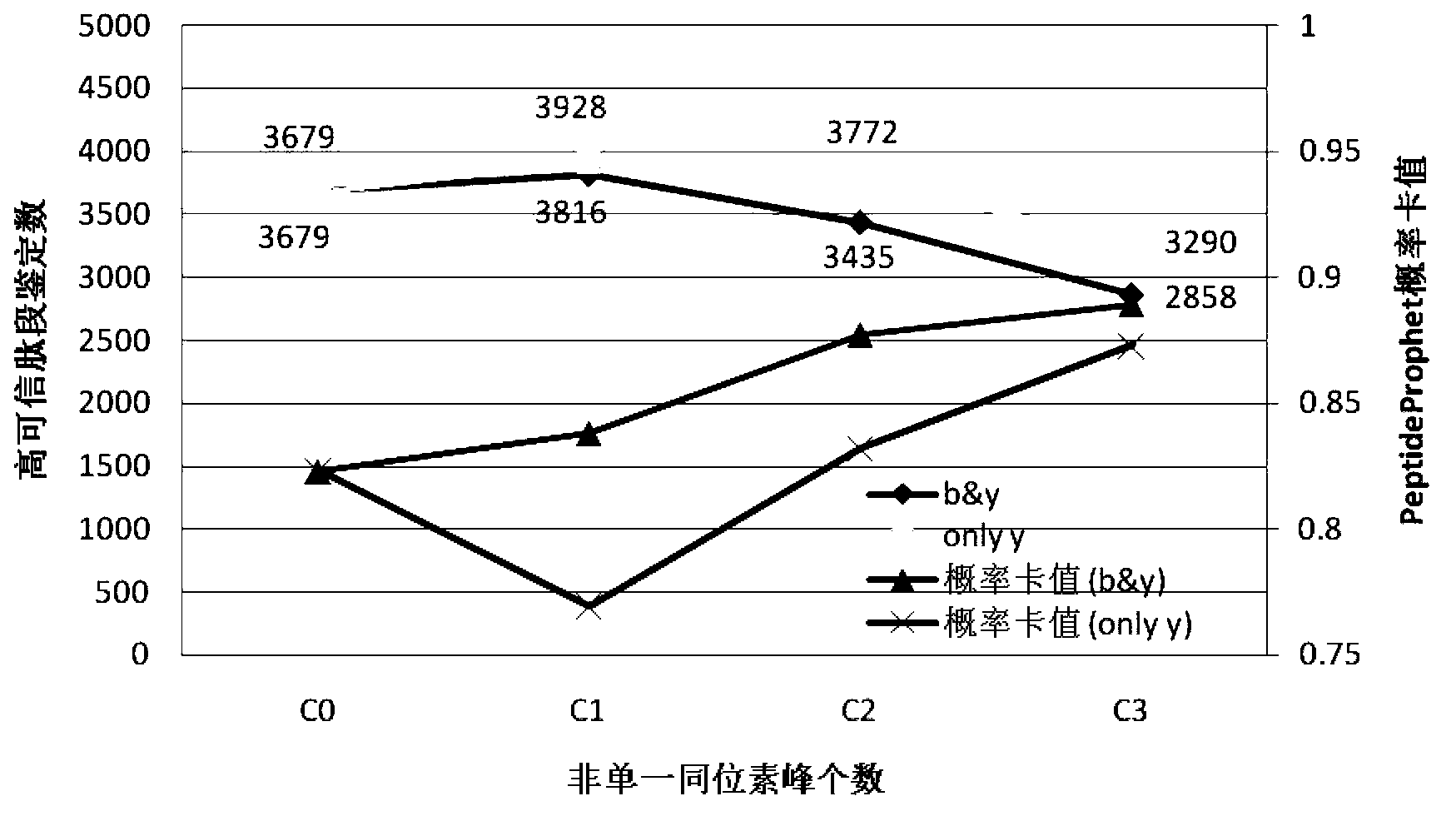
[0069] The HCD secondary mass spectrograms of 16,479 peptides obtained in 2 of Example 1 were processed according to the basic method of Step 3 and Step 4 of Example 1 as follows:
[0070] C1 is the first isotope peak added, and "only y" is the isotope peak with only y ions added. "only y"+C1 is the best condition, that is, only the first isotope peak of y ion is added. C0 means no isotopic peaks added.
[0071] A, "b&y"+C0 treatment group (for the control group in Example 1)
[0072] B. "b&y"+C1 treatment group
[0073] B.1) Format conversion: the method is the same as step 3 of embodiment 1;
[0074] B.2) Search, generate theoretical spectrum and match scoring: basically the same as Step 4 of Example 1, the difference is that in the theoretical spectrum, except for the mass-to-charge ratio of the b and y ion monoisotope peaks of candidate peptides and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com