Group of aptamers for specific recognition of beta-bungatotoxin and use thereof
A technology of bungarotoxin and nucleic acid aptamer, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., and can solve problems such as poor stability, insufficient adsorption of enzyme-labeled plates, and weak affinity , to achieve the effects of small molecular weight, easy chemical modification and labeling, and short cycle time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
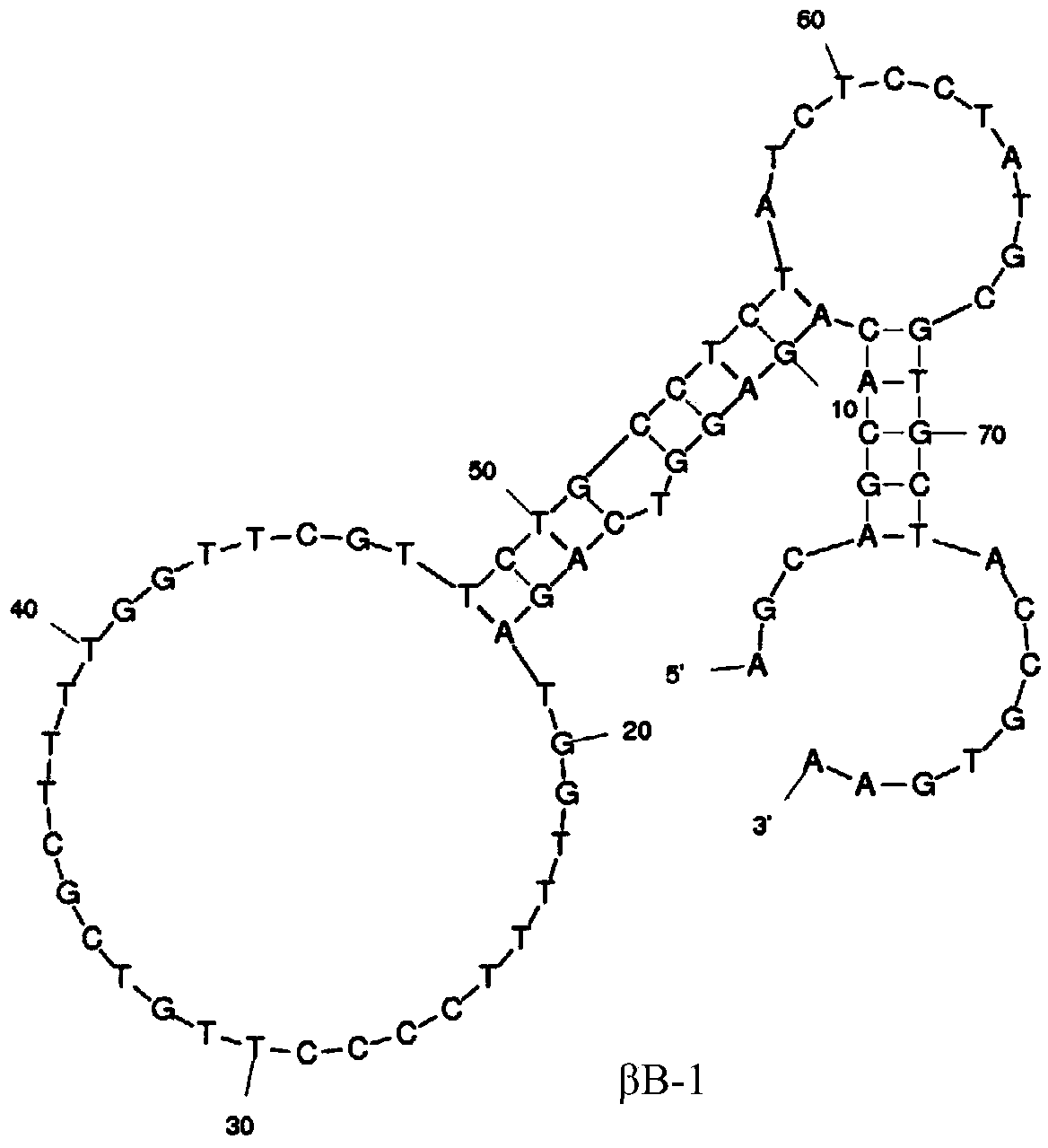
[0032] Example 1 Screening of nucleic acid aptamers specifically binding to β-bungarotoxin
[0033]1. Construct a random single-stranded DNA library and primers: design a random single-stranded DNA library with a fixed sequence of 20 bases at both ends and a random sequence of 40 bases in the middle, namely: 5′-AGC AGC ACA GAG GTC AGA TG -N40-CCT ATG CGT GCT ACC GTG AA-3'. Where N represents any one of bases A, G, C, and T, and N40 represents a random fragment length of 40 bases. Primer 1: 5'-fluorescein-AGC AGC ACA GAG GTC AGA TG-3', Primer 2: 5'-TTC ACG GTA GCA CGC ATA GG-phosphate group-3'. The library and primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd. Binding buffer (20mM HEPES, pH7.4; 150mM NaCl; 5mM KCl; 2mM MgCl 2 ;2mM CaCl 2 ), primers use ddH 2 O was prepared into a 100 μM stock solution and stored at -20°C for later use.
[0034] 2. SELEX screening: Prepare β-bungarotoxin with carbonate buffer (CBS, pH 9.6) to a concentration of 10 μg / mL, t...
Embodiment 2
[0037] Example 2 Determination of the binding force of each round of nucleic acid aptamer library and β-bungarotoxin by digesting fiber membrane method
[0038] Nitrocellulose membranes (HAWP02500nitrocellulose filters, Millipore, 0.22μm) were soaked in 0.5M KOH solution for 30min, then soaked with ddH 2 O was fully washed, then placed in the binding buffer, shaken for 40min, placed in a new binding buffer, and stored at 4°C. Each round of fluorescein-labeled ssDNA library was prepared to a concentration of 1 μM, 10 μL was added to 1 mL of binding buffer, placed at 90 ° C for 10 min, then placed on ice for 10 min, and at room temperature for 10 min. Add 4 μL of 1 mg / mL β-bungarotoxin, incubate at room temperature for 1 h, and control without adding β-bungarotoxin, then filter the membrane and wash the filter membrane with 1 mL of binding buffer. The filtrates were combined, and the fluorescence intensity of the filtrate was measured with a Perkin Elmer LS55 fluorescence spect...
Embodiment 3
[0039] Example 3 Determination of the dissociation constant Kd value of the nucleic acid aptamer by the fluorescence anisotropy method
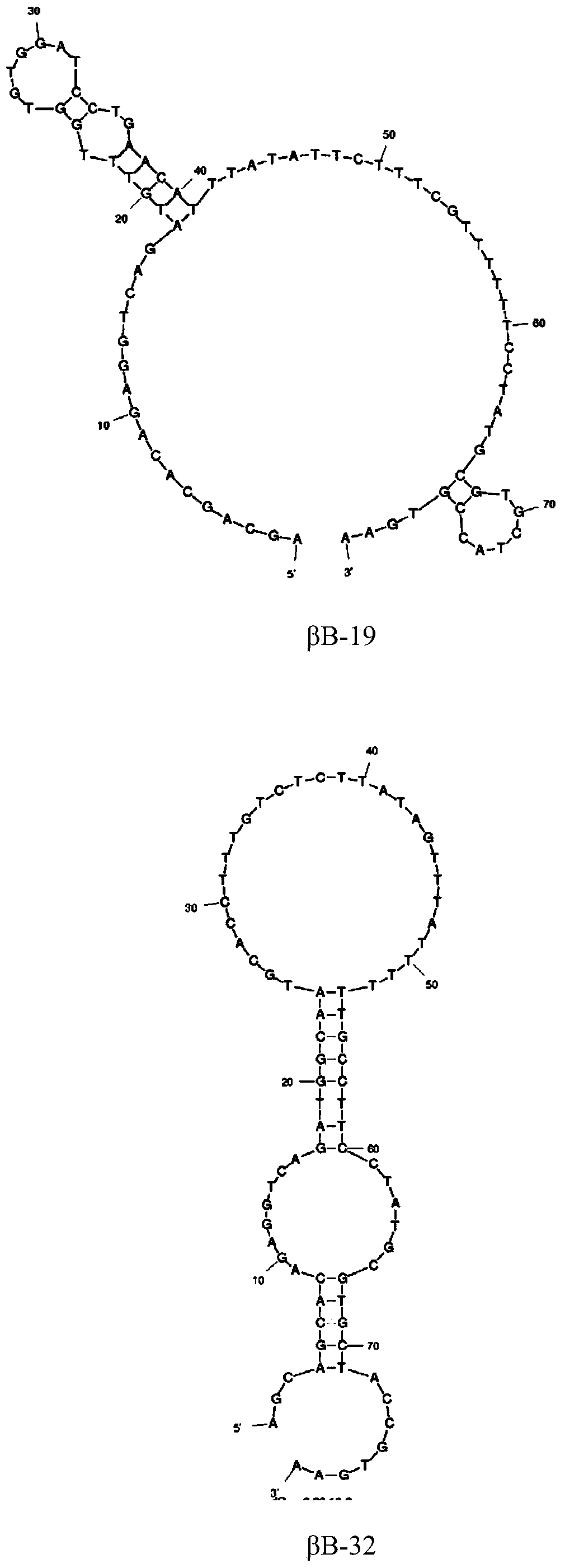
[0040] Fluorescence-labeled aptamers βB-1, βB-19, βB-32, and βB-20 were artificially synthesized, and 4 μL of aptamers (1 μM) were added to 2 mL of binding buffer to measure the fluorescence anisotropy value. Then titrate with different concentrations of β-bungarotoxin, and measure the fluorescence anisotropy value at each concentration. Determination parameters are the same as in Example 2. Use the concentration of β-bungarotoxin as the abscissa, and the increment of the fluorescence anisotropy value as the ordinate, and use SigmaPlot10.0 software to do nonlinear regression to analyze the dissociation constant (Kd) value of the aptamer.
[0041] The Kd values of the aptamers βB-1 and βB-20 were both lower than 100nM, 65.9nM and 83.8nM respectively, and the measurement results were as follows image 3 shown. The Kd value of βB-19 is 540 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com