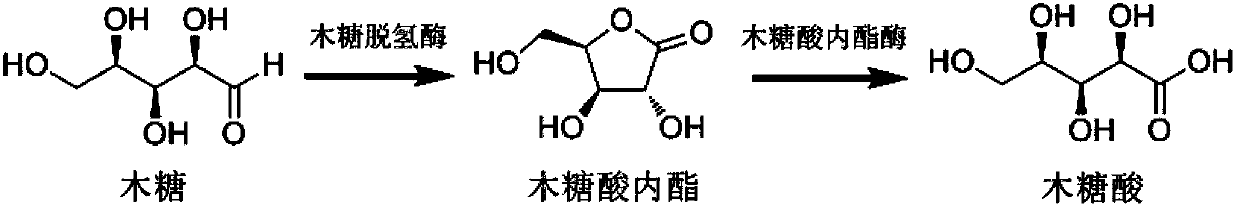
Biological synthesis method of xylosic acid
A technology of xylonic acid and xylose, applied in the field of recombinant engineering acceptor bacteria, can solve problems such as restricting application, restricting commercial application, environmental pollution, etc., and achieves the effects of reducing production cost, reducing oxidation, and avoiding reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
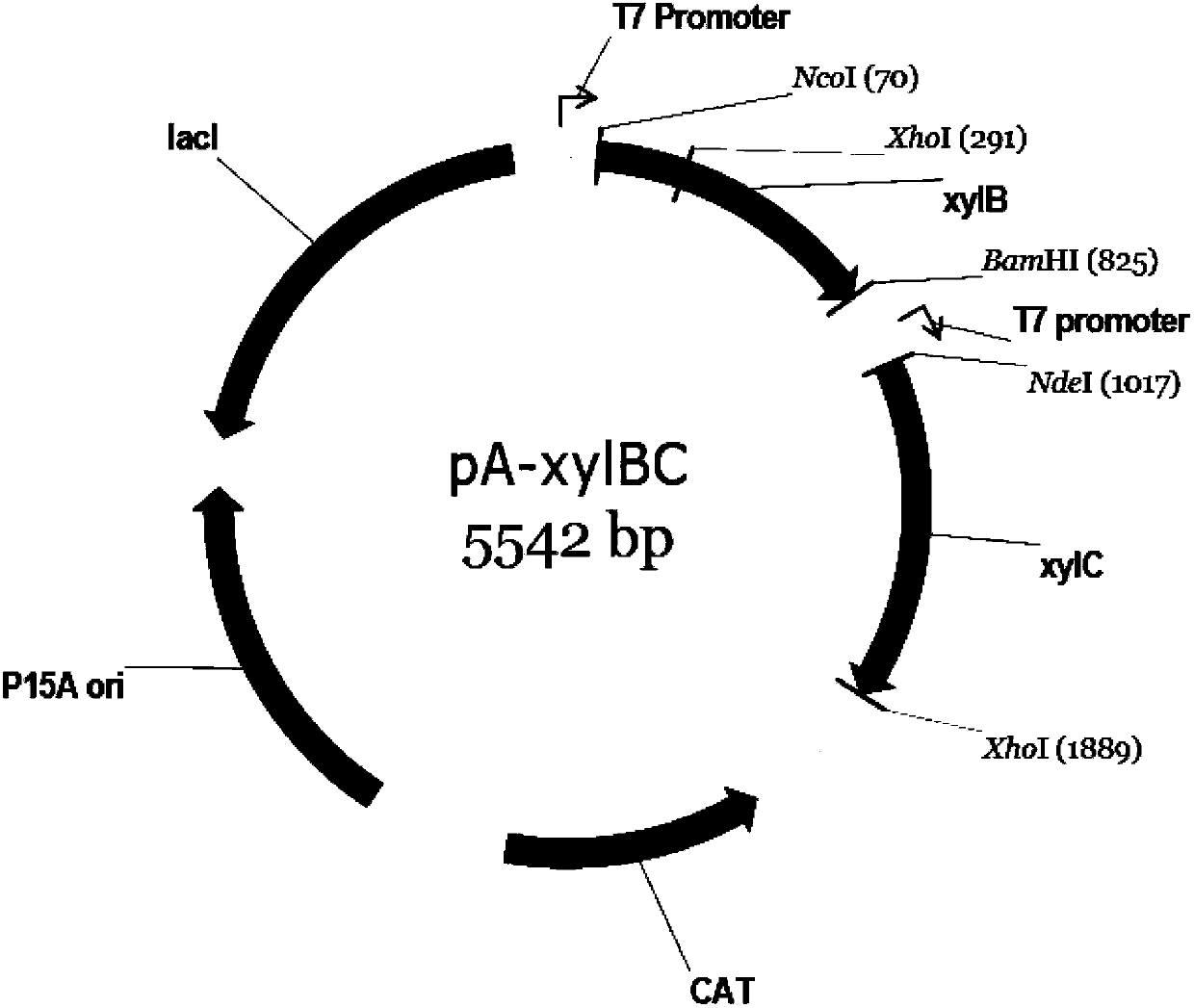
[0058] The construction of the co-expression vector of xylose dehydrogenase gene (xylB) and xylonolactonase gene (xylC) is as follows:
[0059] Using oligonucleotides 5'-CAT GCC ATG GGC ATG TCC TCA GCC ATC TATCCC AG-3' and 5'-CGC GGA TCC TCA ACG CCA GCC GGC GTC GAT C-3' as primers, Genomic DNA of Caulobacter crescentus As a template, the xylose dehydrogenase gene (xylB gene) (SEQ ID No: 1) was amplified by the polymerase chain reaction (PCR) method, and NcoI and BamHI restriction enzymes were introduced at the 5' end and 3' end respectively. site, and then clone the gene into the pACYCduet-1 (purchased from Novagen) vector with the above-mentioned restriction site to obtain the recombinant plasmid pA-xylB;
[0060] Use oligonucleotides 5'-GGG AAT TCC ATA TGA CCG CTC AGG TTA CATGCG-3' and 5'-CCG CTC GAG TTAAAC CAG ACG AAC TTC GTG C-3' as primers, and Genomic DNA of Caulobacter crescentus as template , the xylonolactonase gene (xylC gene) (SEQ ID No: 3) was amplified by the pol...
Embodiment 2
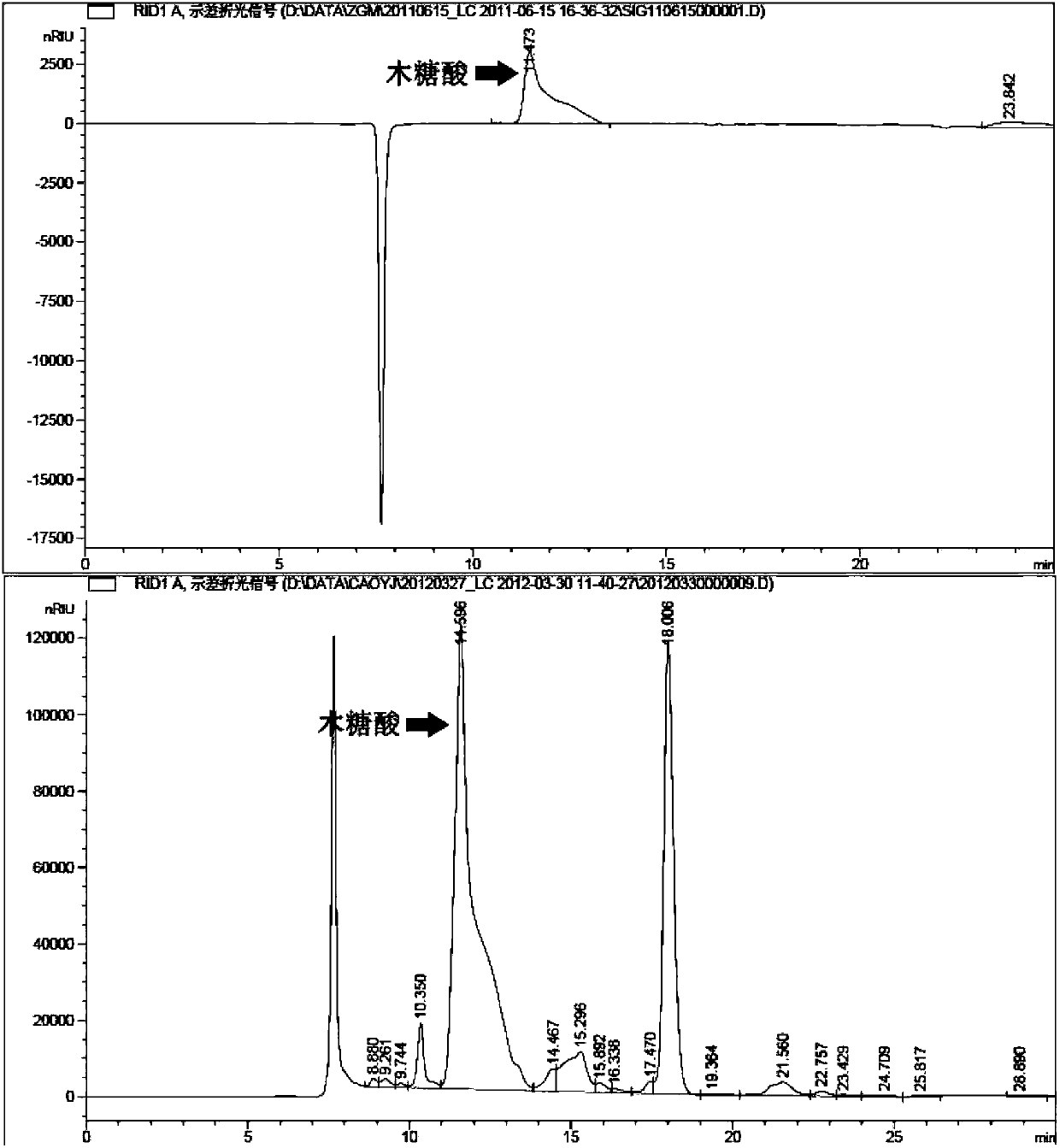
[0062] Preparation of recombinant engineered Escherichia coli strains for synthesizing xylonic acid, and using the strain to ferment and convert D-xylose to generate xylonic acid, the specific process is as follows:
[0063] The recombinant plasmid pA-xylBC constructed in Example 1 was extracted by alkaline lysis, and 10 μl of recombinant plasmid pA-xylBC was transformed into Escherichia coli BL21 (DE3) competent cells by the heat shock transformation method, and then 50 μl of the transformed bacterial solution was applied to Cloth contains 34μg·mL -1 Positive clones were screened on chloramphenicol LB plates, and the grown colonies were recombinant E. coli strains co-expressing xylose dehydrogenase and xylonolactonase. The recombinant E. coli strain containing the recombinant plasmid pA-xylBC can be further verified by sequencing.
[0064] Pick a single colony of recombinant Escherichia coli that has been constructed and inoculate it to a concentration of 34 μg·mL -1 Chlora...
Embodiment 3
[0067] Preparation of recombinant engineered Escherichia coli strains for synthesizing xylonic acid, and using the strain to ferment and convert D-xylose to generate xylonic acid, the specific process is as follows:
[0068] The recombinant plasmid pA-xylBC constructed in Example 1 was extracted by alkaline lysis, and 1 μl of recombinant plasmid pA-xylBC was transformed into Escherichia coli BL21 (DE3) competent cells by the heat shock transformation method, and then 100 μl of transformed bacterial liquid was applied to Cloth contains 34μg·mL -1 Positive clones were screened on chloramphenicol LB plates, and the grown colonies were recombinant E. coli strains co-expressing xylose dehydrogenase and xylonolactonase. The recombinant E. coli strain containing the recombinant plasmid pA-xylBC can be further verified by sequencing.
[0069] Pick a single colony of recombinant Escherichia coli that has been constructed and inoculate it to a concentration of 34 μg·mL -1 Chlorampheni...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com