Method for safely producing 1,3-propanediol
A propylene glycol, safe technology, applied in the field of bioengineering, can solve the problems of pathogenicity concerns, insufficient, low pathogenicity correlation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 The genes contained in VH1 and VH2 are potential disease-causing genes
[0022] Compare the gene sequences in VH1 and VH2 with the existing virus gene bank. The virus gene bank can be queried in the virus gene database collected and organized by the Institute of Pathogen Biology, Chinese Academy of Medical Sciences, and the Key Laboratory of Pathogen System Biology, Ministry of Health ( http: / / www.mgc.ac.cn / VFs / main.htm). The query results show that the genes contained in the large fragment VH1 and VH2, except fes , wxya , entS In addition, other genes have been confirmed to be disease-causing genes by existing studies, and were originally found in Escherichia coli.
[0023]
Embodiment 2
[0024] Embodiment 2, The reported Klebsiella pneumoniae genome contains large fragments of VH1 and VH2 and is highly homologous
[0025] Table 1: Analysis of VH1 fragments in reported Klebsiella genomes
[0026]
[0027] The genetic data of SEQ ID NO 1 to SEQ ID NO 22 were converted into protein data and compared with the Klebsiella genome data reported on NCBI (http: / / www.ncbi.nlm.nih.gov / ) , the specific compared genomes are: NC_017540, NC_011283, NC_016845, NC_018522, NC_012731, NC_009648.
[0028] Table 2: Analysis of VH2 fragments in reported Klebsiella genomes
[0029]
[0030] After alignment, the genes in SEQ ID NO 1 to SEQ ID NO 22 were located on each genome. Specific genetic data search, comparison, and gene location adopt general methods in molecular biology, and will not be described here. The comparison positioning results are shown in Table 1 and Table 2. As can be seen from Table 1 and Table 2, the genomes of each Klebsiella pneumoniae contain VH1 a...
Embodiment 3
[0032] Embodiment 3, VH1 and VH2 gene fragments also exist in 1,3-PD Klebsiella pneumoniae
[0033] The 1,3-PD-producing strain uses ATCC49790. The upstream and downstream primers of VH1 are derived from the gene fepA (SEQ ID NO 1) upstream and gene entA (SEQ ID NO 13). The upstream and downstream primers of VH2 are derived from the gene fimB (SEQ ID NO 14) upstream and gene f (SEQ ID NO 22).
[0034] ATCC49790 was cultured overnight at 37°C in LB medium (0.5% yeast extract, 1% tryptone, 1% NaCl, pH 7.0), and the genome was extracted. Using the extracted ATCC49790 genome as a template, the designed VH1 and VH2 upstream and downstream primers were used for PCR reaction. After the PCR reaction, the target band was recovered and sequenced, and the gene analysis was performed on the target band after sequencing.
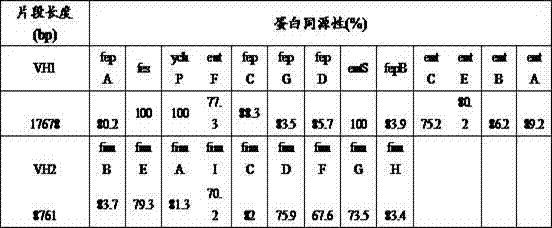
[0035] The analysis results are shown in Table 3. As can be seen from Table 3, the genome of ATCC49790 contains VH1 (17678bp) and VH2 (8761bp) fragments, and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com