A microfluidic chip for nucleic acid sequencing
A microfluidic chip and nucleic acid sequencing technology, applied in laboratory containers, enzymology/microbiology devices, specific-purpose bioreactors/fermenters, etc., can solve problems such as expensive raw materials and complex substrate modification, Achieve the effects of saving reagent consumption, high reaction and washing efficiency, and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
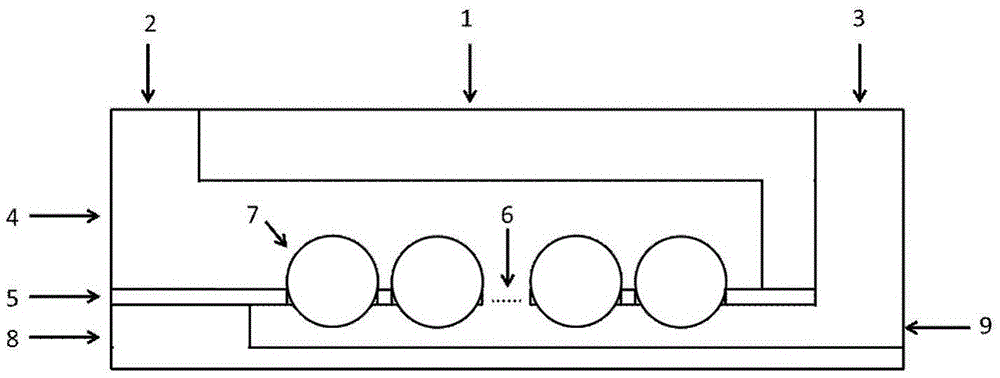
[0041] a) Preparation of the upper layer chip: use a 3cm×7cm quartz plate, cut out three microchannels with a length of 5.2cm, a width of 5mm, and a depth of 50μm in the quartz plate through laser micromachining, and the interval between the three channels is 2mm, and a diameter of 1mm is punched out. Liquid outlet and inlet.
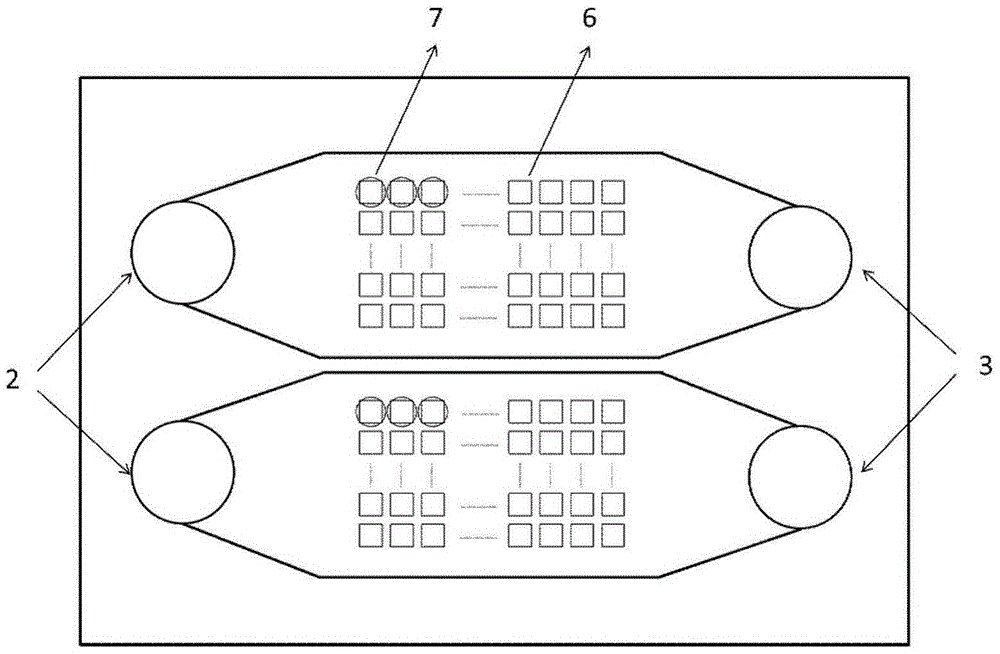
[0042] b) Fabrication of the middle layer chip: use a 3cm×7cm metal nickel film to obtain a square through-hole microarray by electrochemical etching at the position corresponding to the microchannel of the upper chip. The edge of the microwell array is 4 μm from the edge of the channel, and 2 mm from the entrance and exit of the channel.
[0043] c) Preparation of the lower layer chip: using a 3 cm x 7 cm quartz plate, three microchannels with a length of 5.2 cm, a width of 5 mm and a depth of 50 μm were cut out in the quartz plate by laser micromachining.
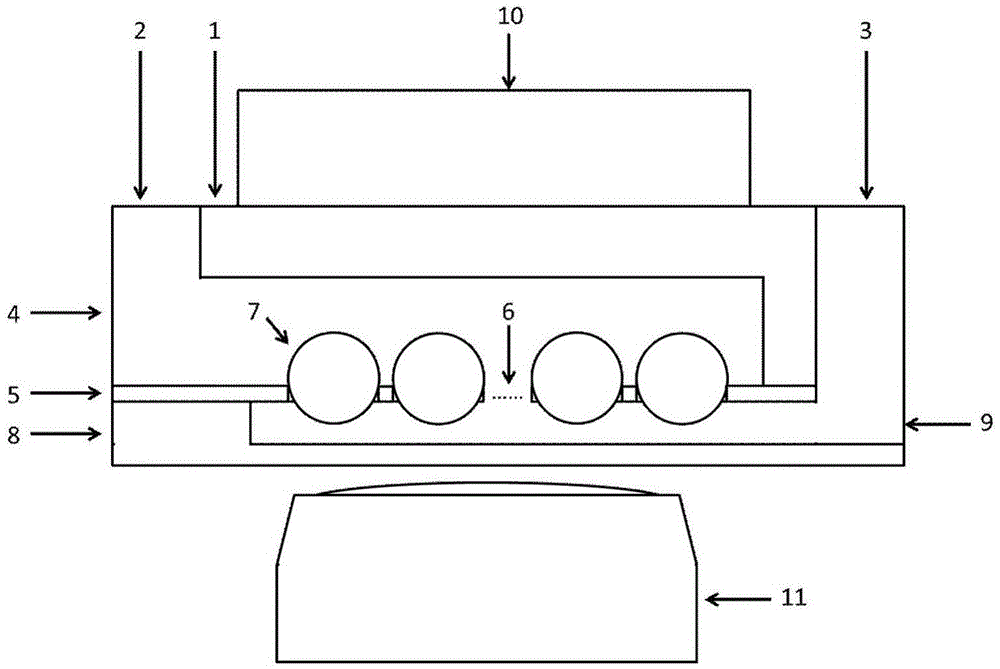
[0044] d) Chip assembly: The three-layer chip is connected together by 5 μm thick glue, and co...
Embodiment 2
[0047] a) Preparation of the upper chip: use a 3cm×7cm silicon wafer, and prepare three microchannels with a length of 5.2cm, a width of 5mm, and a depth of 20μm in the silicon wafer by wet etching. The three channels are separated by 2mm and the diameter is 1mm. liquid outlet and inlet.
[0048] b) Preparation of the middle layer chip: use a 3cm×7cm silicon wafer, and dry-etch a square through-hole microarray at the position corresponding to the microchannel of the upper chip. The edge of the hole array is 4 μm away from the edge of the channel, and 2 mm away from the entrance and exit of the channel.
[0049] c) Preparation of the lower layer chip: using a 3 cm x 7 cm quartz plate, three microchannels with a length of 5.2 cm, a width of 5 mm and a depth of 20 μm were cut out in the quartz plate by laser micromachining.
[0050] d) Chip assembly: The three-layer chip is connected together through a bonding process, and connected to the peripheral pipeline through the liquid ...
Embodiment 3
[0053] a) Preparation of the upper layer chip: use a 3cm×7cm silicon wafer, and print adhesive glue containing 3 microchannels with a length of 5.2cm, a width of 5mm, and a depth of 100μm on the quartz wafer through a screen printing process. The three channels are spaced at 2mm, and Make a liquid outlet and inlet with a diameter of 1mm.
[0054] b) Preparation of the middle layer chip: use a 3cm×7cm silicon wafer, and wet-etch at the position corresponding to the microchannel of the upper chip to obtain a square through-hole microarray. The edge of the microwell array is 4 μm from the edge of the channel, and 2 mm from the entrance and exit of the channel.
[0055] c) Preparation of the lower layer chip: use a 3cm×7cm glass sheet, and print adhesive glue containing 3 microchannels with a length of 5.2cm, a width of 5mm, and a depth of 100μm on the quartz sheet by a screen printing process, and the distance between the three channels is 2mm.
[0056] d) Chip assembly: the thr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com