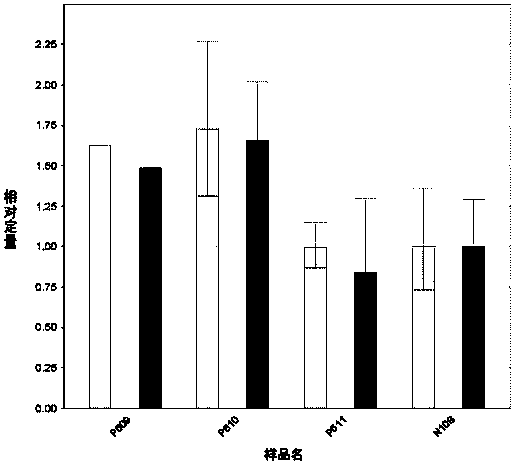
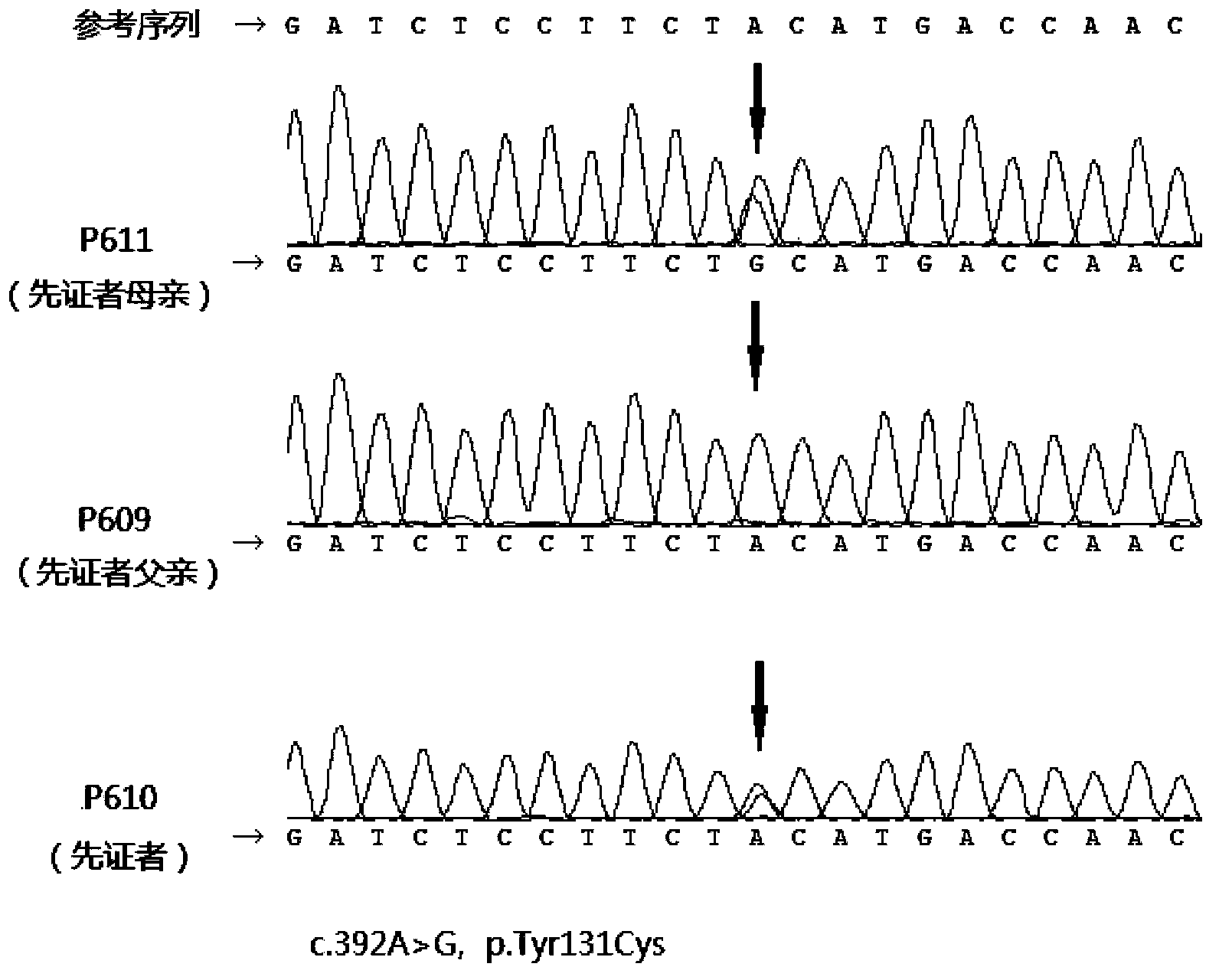
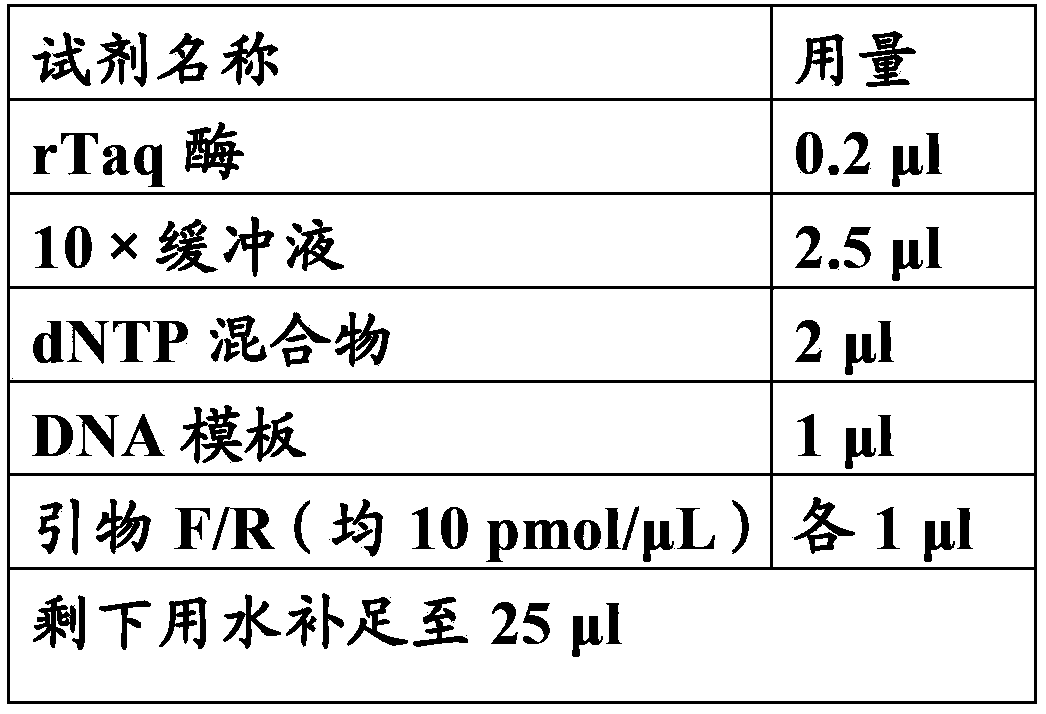
Maple syrup urine disease related gene mutation, and detection method and purpose thereof
A gene, α subunit technology, applied in the field of disease-related mutant genes, can solve problems such as loss, decreased branched-chain α-keto acid decarboxylase activity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] genome extraction
[0068] Peripheral blood was collected from all subjects, and genomic DNA was extracted using the QIAamp DNA BloodMiNi Kit (Qiagen, Hilden, Germany) according to the protocol provided by the manufacturer.
Embodiment 2
[0070] Chip design, library construction, and high-throughput sequencing
[0071] The inventors designed a capture chip (NimblGen, Roche) to capture the exons, splice sites and adjacent intron sequences of the four genes BCKDHA, BCKDHB, DBT and DLD.
[0072] For the method of library construction, please refer to the protocol provided by Illumina (http: / / www.illumina.com / ). The brief description is as follows: Genomic DNA was randomly broken into fragments of 200-300 bp using an ultrasonic instrument (Covaris S2, Massachusetts, USA). Take 1 μg (quantified by Nanodrop) of purified DNA fragments and treat them with T4 DNA polymerase, T4 phosphonucleotide kinase and Klenow fragment of Escherichia coli DNA polymerase to fill in the 5'overhanging ends and remove the 3'overhanging ends. A base is added to the 3' end and then a linker sequence is ligated. After adding adapters, PCR primers containing 8 bp tag sequence (barcode sequence) were used to perform 4 cycles of PCR to obtai...
Embodiment 3
[0074] Variant detection, annotation and comparison with databases
[0075] 1) After the sequencing is completed, use Illumina1.3.4 for image analysis, error rate estimation and base reading to obtain raw data. Low-quality reads (reads) and reads containing adapter contamination are then filtered. We will use BWA to align high-quality reads to the reference genome for reads containing more than 10% of N bases and 50% of the base-occupied reads with a quality value of less than 5 to obtain the only alignment on the genome Read paragraph. Then use SOAPsnp and GATK to detect SNP and indel respectively.
[0076] 2) For the detected SNP and indel variants, we use the internal process to annotate, that is, to determine the genomic environment of the variant and the type of the variant. We focused on non-synonymous mutations, splice acceptor / donor site mutations, and coding region insertion and deletion mutations, the three most likely disease-associated mutations. We compared th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com