Arabidopsis AtPGK2 gene for enhancing salt tolerance of plants and application of arabidopsis AtPGK2 gene
A technology of Arabidopsis thaliana and salt tolerance, applied in the field of plant genetic engineering, can solve problems such as unclear physiological functions, achieve wide application value, and improve salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Arabidopsis Encoding 3-Phosphoglycerate Kinase Gene AtPGK2 clone
[0029] (1) The total RNA of Arabidopsis plants was extracted by the nitrile guanidine-phenol isosulfate method. For the use, preparation and specific operation steps of the drug, refer to the method of Huang Peitang et al. (2002).
[0030] (2) According to known Arabidopsis AtPGK2 Design specific primers for the CDS sequence of the gene and introduce them in the upstream primers Nco I restriction site, introduced in the downstream primer BstE II restriction site.
[0031] Upstream primer: 5'-CATG CCATGG CTTCCACCGCCGCAACTGCA-3' (introduced Nco I restriction site)
[0032] Downstream primer: 5'-G GGTAACC TTAAACAGTGACTGGCGTTGCT-3' (introduced BstE II restriction site)
[0033] (3) Take 1 mg RNA as the template for reverse transcription, with P 2853 For primers using Promega reverse transcriptase ImProm-II TM For reverse transcription, P 2853 The primer sequences, reverse trans...
Embodiment 2
[0049] Embodiment 2: Utilize pCAMBIA1301 vector construction " CaMV 35S - AtPGK2 "Fusion gene
[0050] (1) Extract vector pCAMBIA 1301 plasmid (purchased from CAMBIA Company) from Escherichia coli, use Nco I / BstE Recover large vector fragments after II double enzyme digestion.
[0051] (2) Reclaimed to embodiment 1 AtPGK2 The CDS fragment of the gene is used Nco I / BstE II double enzyme digestion, and the digested fragments were recovered by agarose gel electrophoresis (same as in Example 1).
[0052] (3) Ligate the above two fragments under the catalysis of ligase at 16°C overnight to complete the " CaMV 35S - AtPGK2 "Fusion gene construction.
[0053] Connection system:
[0054] Reagent Amount added (μl) AtPGK2 CDS fragment of the gene (50 ng μl -1 ) 2.0 Large fragment of pCAMBIA1301 vector (50 ng μl -1 ) 3.0 Solution I Ligase 5.0
[0055] (4) Transform Escherichia coli DH5α competent cells with the ligation mixture...
Embodiment 3
[0061] Embodiment 3: Preparation of transgenic Arabidopsis plants
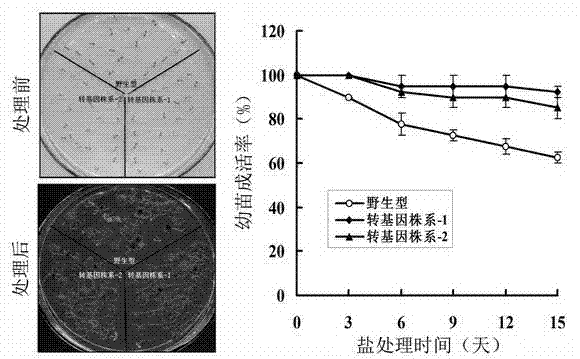
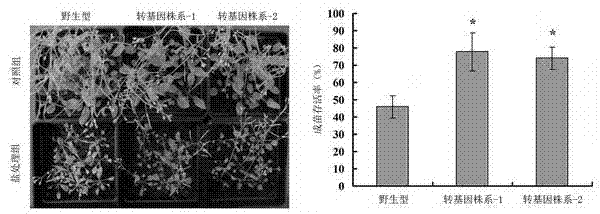
[0062] (1) " CaMV 35S - AtPGK2 "The fusion gene was transformed into Arabidopsis thaliana. The specific transformation method used the Floral dip method mediated by Agrobacterium (Clough and Bent, 1998). The obtained seeds were subjected to 50 mg l -1 For hygromycin resistance screening, normal growing plants were transferred to soil for culture.
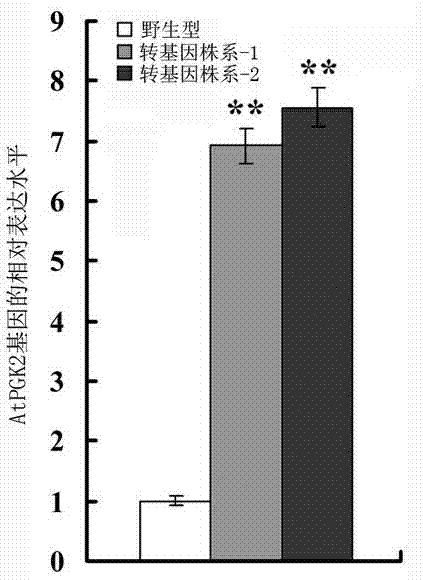
[0063] (2) Real-time quantitative RT-PCR detection of transgenic plants: the transgenic Arabidopsis thaliana identified by PCR in Example 2 was multiplied, and after 50 mg l -1 Hygromycin resistance screened for T 3 To generate homozygous transgenic lines, obtain wild-type and transgenic Arabidopsis seedlings grown for 1 week respectively, extract the total RNA of plant tissue according to the method of Example 1 and reverse transcribe it into mRNA, use the following primers, reaction procedures and reaction System for PCR reaction:
[0064] AtPGK2 Gene detec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com