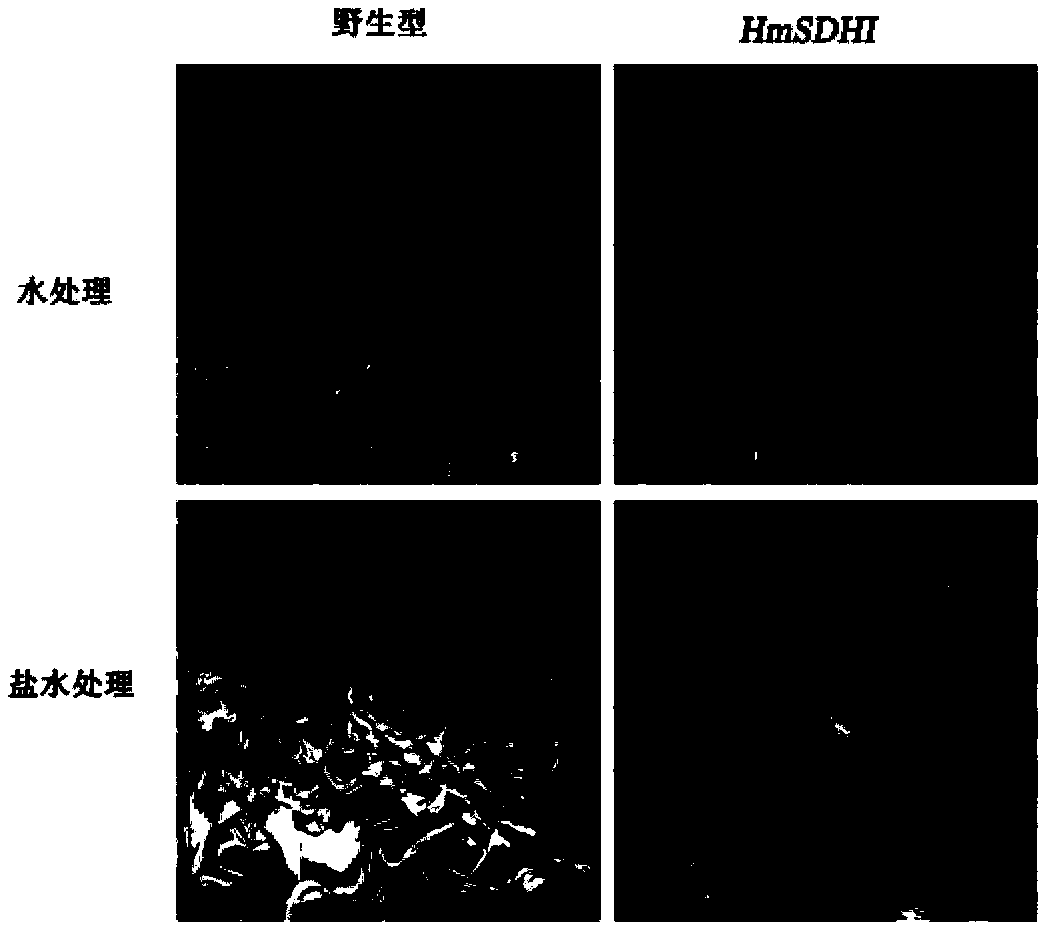
Sorbitol dehydrogenase gene derived from haloarcula marismortui and applications thereof
A technology of sorbitol dehydrogenase and Dead Sea salt, applied in application, genetic engineering, plant genetic improvement, etc., can solve problems such as SDH activity decline
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1 The artificial synthesis of sorbitol dehydrogenase gene HmSDHI
[0034] According to the PTDS (PCR-based two-step DNA synthesis, PTDS) method, the sorbitol dehydrogenase gene derived from Halobacterium dead sea was cloned and chemically synthesized. On the basis of keeping the amino acid sequence of the sorbitol dehydrogenase gene unchanged, Primers were designed to synthesize the sorbitol dehydrogenase gene (HmSDHI) of the present invention. The designed primers are as follows:
[0035] 1.HMSDHI-1:Tm=54,60mer
[0036] GGA,TCC,ATG,AGA,CCA,TCT,GCT,TCT,GTT,GGT,CAG,AGA,CCA,GAA,GAG,AGA,CTC,AAT,CCA,CAG
[0037] 2. HMSDHI-2:Tm=54,60mer
[0038] TGA,CAC,CAA,TGA,CTT,TCA,TGC,CTG,GAA,GAG,AGT,GAG,AGC,ACT,GTG,GAT,TGA,GTC,TCT,CTT
[0039] 3.HMSDHI-3:Tm=54,60mer
[0040] CAT,GAA,AGT,CAT,TGG,TGT,CAC,CAG,AGA,TGA,TGA,TGG,TCC,ACA,ACT,CTT,GGA,GAG,AGA,GAG
[0041] 4.HMSDHI-4:Tm=54,60mer
[0042] GAG,GGT,TCT,GAC,AAG,GGC,TTC,ACC,TGG,ATC,TGG,AGA,TGG,TCT,CTC,TCT,CTC,CAA,GAG...
Embodiment 2
[0095] Example 2 Construction of HMSDHI Agrobacterium Binary Vector
[0096] The ORF of the above-mentioned artificially synthesized HMSDHI gene was amplified by PCR, and the head and the tail were respectively added to Bam HI and Sac I cutting sites, and the DNA fragments recovered were connected with the corresponding enzyme-cut vectors after digestion with Bam HI and Sac I, and identified by enzyme digestion And the correct binary vector pYM6854 was obtained after sequencing.
Embodiment 3
[0097] Embodiment 3 Transformation of Agrobacterium by electric shock method
[0098] 1) Prepare Agrobacterium GV3101 competent, the method refers to MicroPulser TMElectroporation Apparatus Operating Instructions and Application Guide (BIO-RAD Corporation) (Raineri et al., 1990).
[0099] 2) Take 50 μL of GV3101 competent cells, add 1 μL of DNA, and transfer to a 0.2 cm electric shock cup for transformation (400Ω, 2.5KV, 25 μf). Add 1 mL of LB medium containing 1% mannitol to recover the culture for 2 hours (28°C, 250rpm). Take 10 μL and 100 μL respectively and smear LB plates (rifampicin 50 μg / mL, gentamicin 50 μg / mL, chloramphenicol 100 μg / mL).
[0100] 3) Pick several clones from the bacteria grown on the LB plate in step 2, extract the Agrobacterium plasmid by alkaline method, identify by enzyme digestion, and detect by PCR.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com