Kit for magnetic bead method for bacterial genome DNA extraction and extraction method thereof
A genome and kit technology, applied in the field of molecular biology, can solve the problems of high price, insufficient interaction between magnetic beads and buffer, and achieve the effects of stable and reliable quality, increased extraction efficiency, and high purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
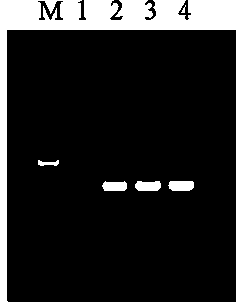
Embodiment 1
[0030] Embodiment 1, monodisperse Fe 3 O 4 SiO 2 - Preparation of AEAPS nanomagnetic beads.
[0031] (1) Monodisperse iron ferric oxide nanospheres were prepared by solvothermal method. The specific preparation process was as follows: Weighed 2.0 g of sodium acetate and 0.25 g of ferric chloride hexahydrate, respectively, and added them to 50 ml of ethylene glycol solution. After magnetic stirring for 1 hour, the system was transferred to a reaction kettle, and reacted at 100° C. for 10 hours. After the reaction was completed, the resulting black solution was centrifuged, and washed 3 times with deionized water and ethanol successively to obtain monodisperse ferric oxide nanospheres (Fe 3 o 4 ), the resulting product was dried in a 60°C oven for use;
[0032](2) Monodisperse silica-coated Fe3O4 nanospheres were prepared by the improved Stäber method. The specific preparation process was as follows: 0.1 g of Fe3O4 nanospheres prepared in step (1) were weighed ...
Embodiment 2
[0034] Embodiment 2: application embodiment.
[0035] (1) Take 1ml of fresh Shigella culture solution and place it in a 2.0ml EP tube, centrifuge at 12000 rpm for 1min, collect the bacteria, add 200ul buffer A, beat and mix well, and resuspend the bacteria;
[0036] (2) Add 200ul buffer B to the EP tube in step (1), incubate in a 56°C water bath for 10min;
[0037] (3) Add 200ul of buffer C to the EP tube incubated in step (2), beat and mix, and let stand at room temperature for 3 minutes;
[0038] (4) Add 30ul of magnetic bead suspension D to the EP tube after step (3), beat and mix well, place the EP tube on the magnetic stand, let it stand for 30s, and carefully remove the liquid when the magnetic beads are completely absorbed;
[0039] (5) Remove the EP tube in step (4) from the magnetic stand, add 500ul of buffer E, beat and mix, place the EP tube on the magnetic stand, let it stand for 30s, and carefully remove the liquid when the magnetic beads are completely absorbe...
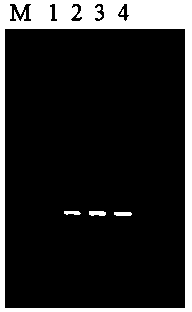
Embodiment 3
[0045] Embodiment 3: application embodiment.
[0046] (1) Take 1ml of fresh Staphylococcus aureus culture solution and place it in a 2.0ml EP tube, centrifuge at 12000 rpm for 1min, collect the bacteria, add 200ul buffer A, 20μl lysozyme (20mg / ml), vortex and mix well, and EP Tubes were incubated in a 37°C water bath for 1 hour, and mixed 3 times during the period;
[0047] (2) Add 200ul buffer B to the EP tube in step (1), incubate in a 56°C water bath for 10min;
[0048] (3) Add 200ul of buffer C to the EP tube incubated in step (2), beat and mix, and let stand at room temperature for 3 minutes;
[0049] (4) Add 30ul magnetic bead suspension D to the EP tube after step (3), place the EP tube on the magnetic stand, let it stand for 30s, and carefully remove the liquid when the magnetic beads are completely absorbed;
[0050] (5) Remove the EP tube in step (4) from the magnetic stand, add 500ul of buffer E, beat and mix, place the EP tube on the magnetic stand, let it stan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com