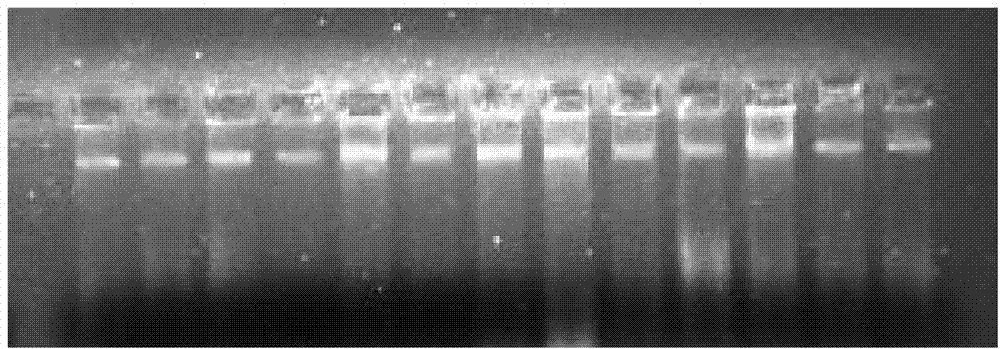
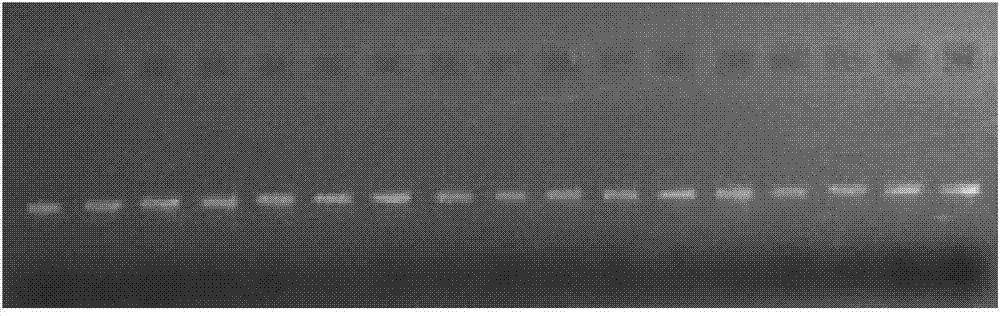
Molecular method for quickly identifying closely related species of juniperus
A technology for the genus and species of Juniper, applied in the molecular field of rapid identification of related species of Juniper, can solve the problem of slow mutation rate, difficulty in accurately identifying and identifying related species of Juniper, and inability to accurately distinguish relatives of gymnosperms species, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] A molecular method for quickly identifying related species of Juniper, including the following steps:
[0080] 1. For the collected dried leaves or seeds of Juniper species, use CTAB method to extract total leaf DNA or seed endosperm DNA; the specific procedures are as follows:
[0081] 1) Take a mature seed of a naturally dried juniper species, break the outer seed coat; infuse the seed with distilled water for 10-24 hours, separate the endosperm of the haploid in the seed with tweezers, and store it in a refrigerator at 4°C;
[0082] 2) Grind the endosperm or crush the leaves: For the separated haploid endosperm, put it in a pre-cooled mortar, put the endosperm of a seed together, add an appropriate amount of polyvinylpyrrolidone (PVP) powder and liquid nitrogen, quickly Grind into fine powder; for dry blades, directly weigh 0.02g with an electronic balance, then put it into a 2.0mL centrifuge tube together with stainless steel beads, and add an appropriate amount of PVP pow...
Embodiment 2
[0135] The present invention is used for rapid molecular identification of related species of Juniper: the collected leaves or seeds of related species of Juniper: Qilian Juniper, Dense Juniper, Little Juniper, Square Juniper and Tibetan Juniper, Perform DNA extraction, and use the nuclear gene primer Chi1 provided by the present invention to perform PCR amplification and sequencing of DNA samples, compare and analyze the differences between the tested sequence and the sequence of the present invention, and construct a phylogenetic tree of all sequences And cluster analysis to carry out molecular identification of Juniper species;
[0136] A molecular method for quickly identifying related species of Juniper, including the following steps:
[0137] 1. Extract the DNA of different individuals of Juniper genus Qilian Juniper, Dense Juniper, Little Juniper, Square Juniper and Tibetan Juniper;
[0138] Steps 1 to 6 are the same as steps 1 to 6 of the first embodiment;
[0139] Seven, the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com