Method for constructing Copepoda full-length cDNA (complementary deoxyribonucleic acid) library
A copepod and library technology, which is applied in the field of constructing a copepod full-length cDNA library, can solve the problems of long time consumption, low RNA content, complicated steps and the like, and achieves the effects of short time consumption, small sample volume and simple method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
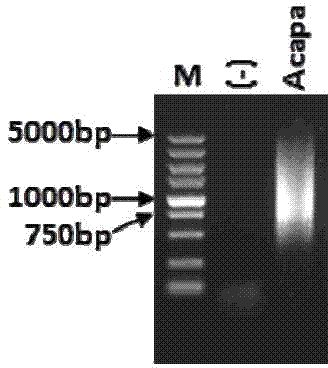
Image
Examples
Embodiment 1
[0045] Using the method above, the full-length cDNA file of Daphnia pacificis was constructed quickly and efficiently.
[0046] library, and the library was analyzed based on the NCBI database and Blast2Go software. Concrete steps
[0047] The steps are as follows:
[0048] (1) Copepod RNA extraction:
[0049] To extract high-quality copepod total RNA, the steps are briefly described as follows:
[0050] (1) The sample is fixed. Copepod samples were fixed using Trizol reagent in 1.5 ml centrifuge tubes (a).
[0051] (2) Sample homogenization. Transfer most of the Trizol in tube (a) to centrifuge tube (b) and homogenize the sample with a grinding rod. Transfer the Trizol back to tube (a), rinse the grinding rod during the transfer, mix well and centrifuge, and transfer the supernatant to tube (b). Grind again 1-2 times.
[0052](3) Transfer the supernatant back to tube (a), add 200 μl chloroform, vortex mix for 1 min, and centrifuge at 10,000 g at 4oC for 15 min.
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com