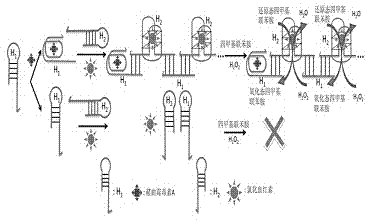
Method for constructing alkaline/surfactant/polymer compound system detecting probe
A ternary composite, detection probe technology, applied in the field of biosensing, to achieve the effects of improving detection sensitivity, reducing costs, and avoiding molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
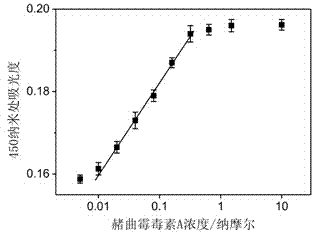
[0024] Add 10 μL of DNA H to 50 μL of a certain concentration of ochratoxin A solution 1 , (H 1 The DNA sequence is GATCGGGTGT GGGTGGCGTA AAGGGAGCAT CGGACACGCC ACCCACAC) aqueous solution, so that H 1 The final concentration of 1nM, then add 10μL CaCl 2 solution and 10 μL KCl solution to make CaCl 2 The final concentration of KCl was 10 mM, and the final concentration of KCl was 20 mM, and then 10 μL of hemin solution was added to make the concentration of hemin 150 nM, mixed evenly, and reacted for 10 minutes. Then add 10 μL of another DNA, namely H 2 (H 2 The DNA sequence is CCACACCCGA TCCTGGGAGG GAGGGAGGGG TGTGGGTGGC G) aqueous solution, so that H 2 The final concentration was 1nM, mixed evenly, and reacted for 1 hour. Then add 80 μL containing H 2 o 2 Tetramethylbenzidine solution, (after adding the H in the solution 2 o 2 Concentration: 0.03%, tetramethylbenzidine concentration: 0.2 μg / mL) and mix evenly, react for 1 hour, then add 50 μL of 1M hydrochloric acid, ...
Embodiment 2
[0027] Add 10 μL of DNA H to 50 μL of a certain concentration of ochratoxin A solution 1 , (H 1 The DNA sequence is GATCGGGTGT GGGTGGCGTA AAGGGAGCAT CGGACACGCC ACCCACAC) aqueous solution, so that H 1 The final concentration of 50nM, then add 10μL CaCl 2 solution and 10 μL KCl solution to make CaCl 2 The final concentration of KCl was 10 mM, and the final concentration of KCl was 20 mM, and then 10 μL of hemin solution was added to make the concentration of hemin 150 nM, mixed evenly, and reacted for 10 minutes. Then add 10 μL of another DNA, namely H 2 (H 2 The DNA sequence is CCACACCCGA TCCTGGGAGG GAGGGAGGGG TGTGGGTGGC G) aqueous solution, so that H 2 The final concentration is 50nM, mix well, and react for 1 hour. Then add 80 μL containing H 2 o 2 Tetramethylbenzidine solution, (after adding the H in the solution 2 o 2 Concentration: 0.03%, tetramethylbenzidine concentration: 0.2 μg / mL) and mix evenly, react for 1 hour, then add 50 μL of 1M hydrochloric acid, measu...
Embodiment 3
[0030] Add 10 μL of DNA H to 50 μL of a certain concentration of ochratoxin A solution 1 , (H 1 The DNA sequence is GATCGGGTGT GGGTGGCGTA AAGGGAGCAT CGGACACGCC ACCCACAC) aqueous solution, so that H 1 The final concentration is 100nM, then add 10μL CaCl 2 solution and 10 μL KCl solution to make CaCl 2 The final concentration of KCl was 10 mM, and the final concentration of KCl was 20 mM, and then 10 μL of hemin solution was added to make the concentration of hemin 150 nM, mixed evenly, and reacted for 10 minutes. Then add 10 μL of another DNA, namely H 2 (H 2 The DNA sequence is CCACACCCGA TCCTGGGAGG GAGGGAGGGG TGTGGGTGGC G) aqueous solution, so that H 2 The final concentration is 100nM, mix well, and react for 1 hour. Then add 80 μL containing H 2 o 2 Tetramethylbenzidine solution, (after adding the H in the solution 2 o 2 Concentration: 0.03%, tetramethylbenzidine concentration: 0.2 μg / mL) and mix evenly, react for 1 hour, then add 50 μL of 1M hydrochloric acid, mea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com