Second-generation sequencing technology based microbe unicell transcriptome analysis method
A next-generation sequencing technology and transcriptome analysis technology, which is applied in the field of microbial single-cell transcriptome analysis based on next-generation sequencing technology, and can solve problems such as the inability to reveal metabolic function gene expression kinetic data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] In this embodiment, the prokaryotic microorganism cyanobacteria Synechocystis sp. PCC6803 was selected as the experimental material. The transcriptome of the cells (24 and 72 hours of nitrogen starvation) and the transcriptome of the population cells at the corresponding time were analyzed. A microbial single-cell transcriptome analysis method based on next-generation sequencing technology, comprising the following steps:
[0030] (1) Separation of microbial single cells:
[0031] The fresh microbial cyanobacteria Synechocystis sp. PCC6803 cell culture samples were collected by centrifugation at room temperature, immediately suspended in the RNA protection agent RNALater solution and fully resuspended; using NARISHIGE IM-9C microinjection system (Narishige, Tokyo , JP) Under the Olympus IX71 inverted microscope, single cells were selected and isolated, and the isolated microbial single cells were resuspended in RNALater solution;
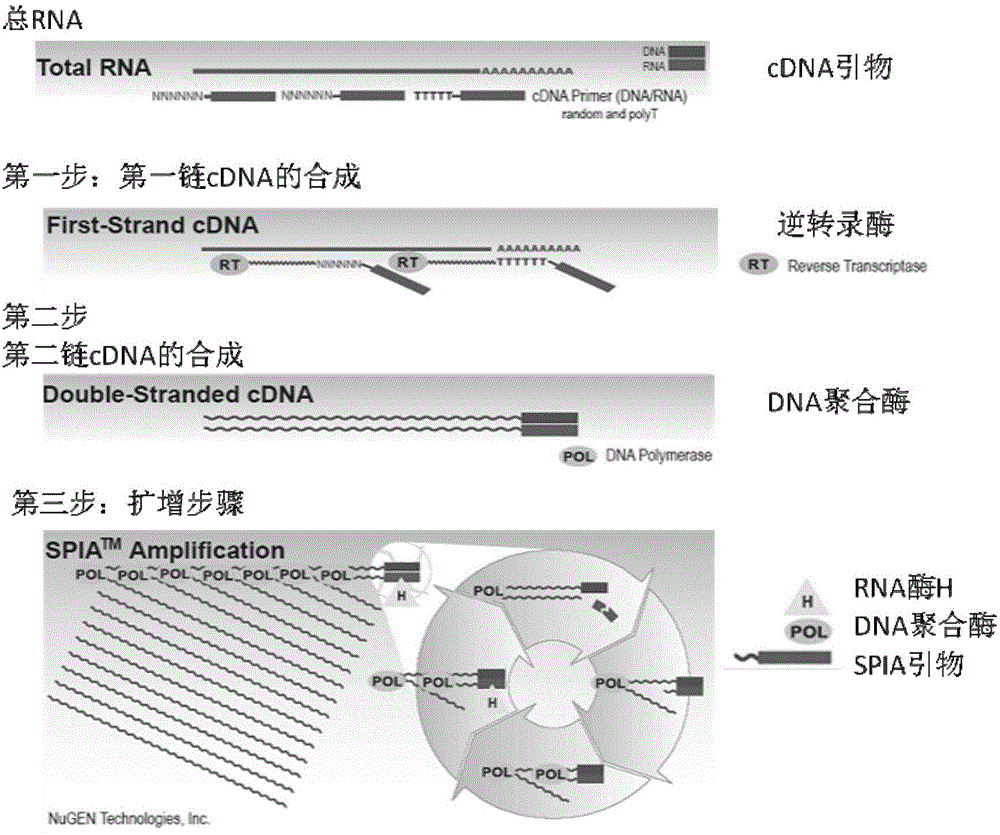
[0032] (2) Extraction of total RNA: ...
Embodiment 2
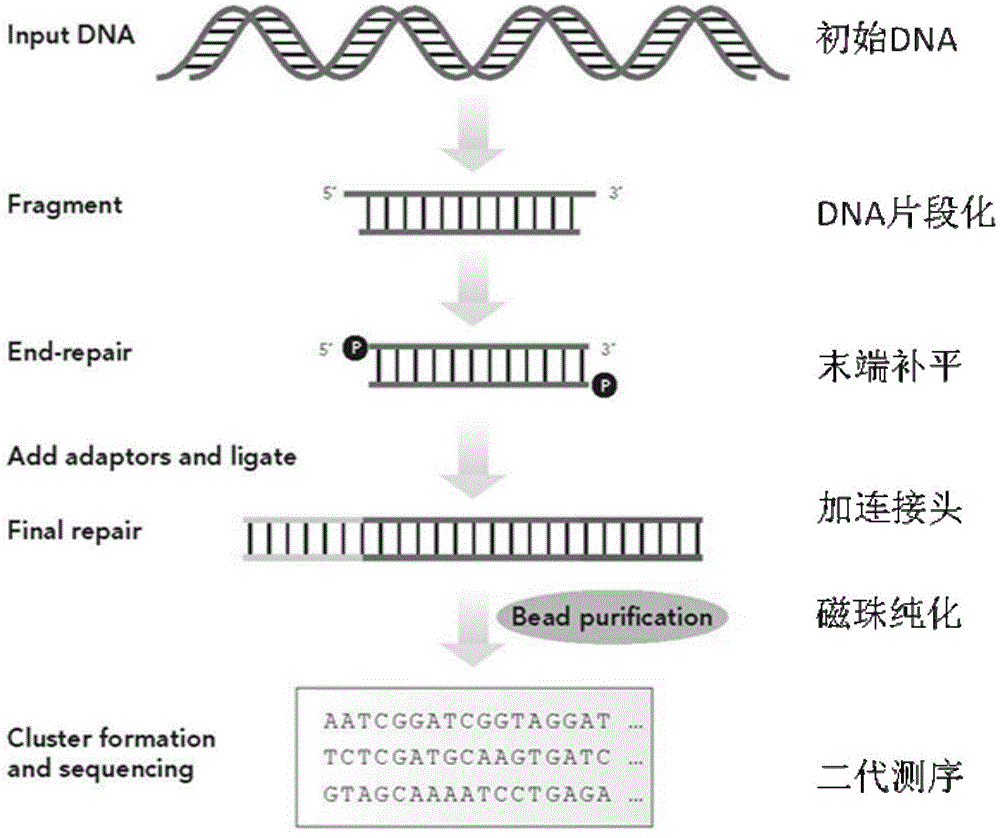
[0058] Quality Control of cDNA Libraries
[0059] Ligate, transform, and clone the cDNA library obtained in step (3) of Example 1, and randomly select clones for sequencing:
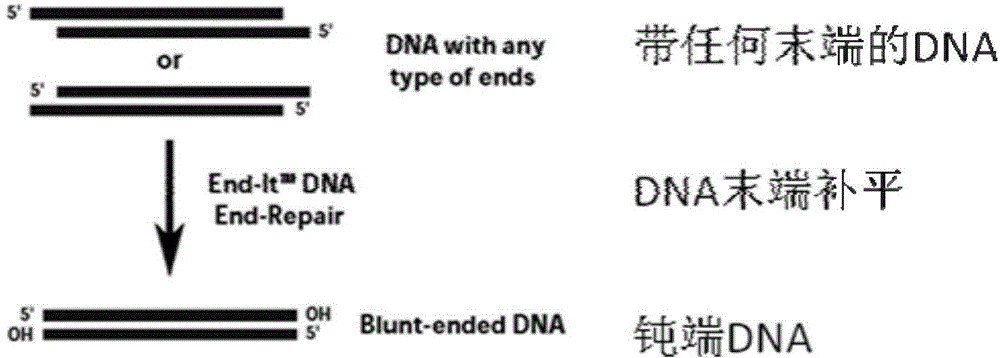
[0060] 1) Use the End-It DNA End-Repair Kit kit from Epicentre, USA to complete the end of the cDNA library, and then use CLONESMART Blunt Cloning Kits to combine the purified cDNA library with 10X End-Repair Buffer, dNTP, ATP, End-Repair Mix the Enzyme Mix evenly, incubate at room temperature for 45 minutes, then incubate at 70°C for 10 minutes to terminate the reaction.
[0061] 2) Use CLONESMART Blunt Cloning Kits to mix the end-filled cDNA library with 4X CloneSmart VectorPremix and CloneSmart DNA Ligase, incubate at room temperature for 30 minutes, then transform into Escherichia coli to construct a clone library, and randomly select single clones from the library for sequencing analyze.
Embodiment 3
[0063] Quantification of RNA-Seq Libraries
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com