A kind of primer and quantitative detection method for detecting Neosphingosine resiniferous bacteria
A quantitative detection method and technology of sphingosine bacteria, which are applied in the field of primers and quantitative detection for the detection of neosphingosine bacterium, can solve the problem of heavy workload, limited research and application of sphingosine resiniferous bacteria, and insufficient detection quantity. Accuracy and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1, primer design and specificity verification
[0059] 1. Primer sequence design
[0060] According to the differences in the conserved sequences of the 16S region of the rDNA of Neosphingobacterium neosphingobacterium and its close relatives registered in GenBank, and according to the principles of primer design, primers were designed using the primer design software Premier 5.0. Then, the primer fragment sequences were compared with the primer specificity in the NCBI database, and a pair of primers NF and NR were screened out. According to the PCR amplification effect and primer amplification specificity of this pair of primers, this pair of primers was selected for the establishment of a molecular detection technology system for Novosphingobium resinovorum.
[0061] 2. Primer specificity verification
[0062] Respectively, Novosphingobium resinovorum (CGMCC 1.3745), underground neosphingobium (Novosphingobium subterraneum, CGMCC1.3516), extracellular polyme...
Embodiment 2
[0069] Embodiment 2, construction and sample analysis of N. resiniferans plasmid standard
[0070] The bacterial DNA extraction kit (DP302-02) from TIANGEN Company was used to extract the DNA of Neosphingobacterium resiniferans. The extracted DNA was amplified by PCR using the specific primer NF / NR of N.
[0071] The amplification reaction system is 50 μL, containing 5 μL of 10×PCR buffer, 2.5mM Mg 2+ 4 μL, 4 μL of dNTPs (2.5 mM each), 1 μL of upstream and downstream primers (10 mM), 2 U of Taq DNA polymerase.
[0072] The amplification reaction program was: pre-denaturation at 94°C for 5 min, deformation at 94°C for 1 min, annealing at 62°C for 30 s, extension at 72°C for 40 s, a total of 30 cycles, and finally extension at 72°C for 10 min.
[0073] The PCR amplified products were detected by 1% agarose gel electrophoresis, and purified by TIANGEN gel recovery kit, and the purified products were connected to the PMD18T vector of Takara Company overnight at 16°C. The ligate...
Embodiment 3
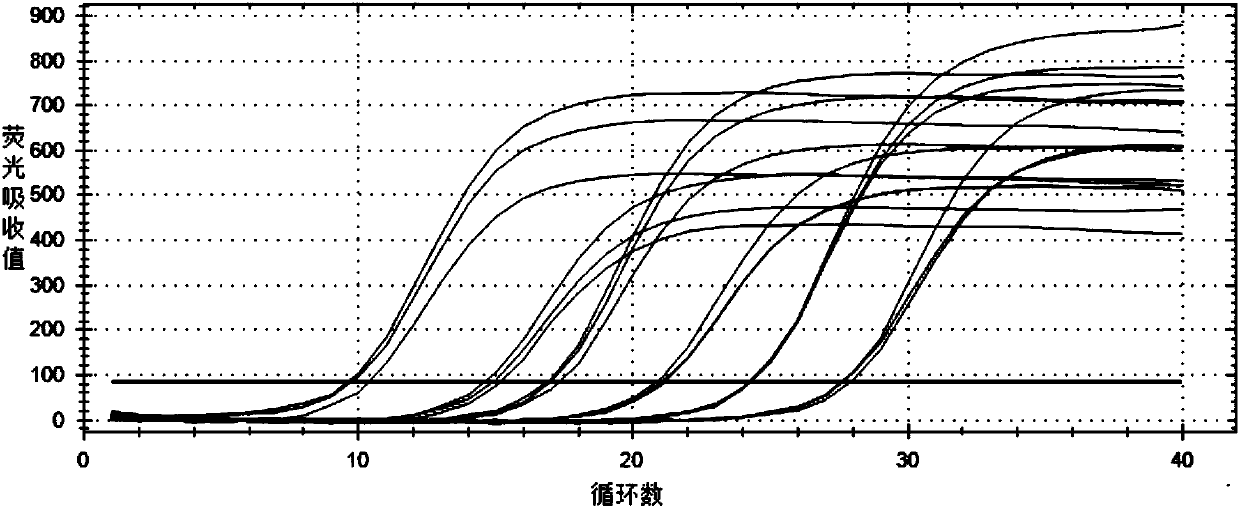
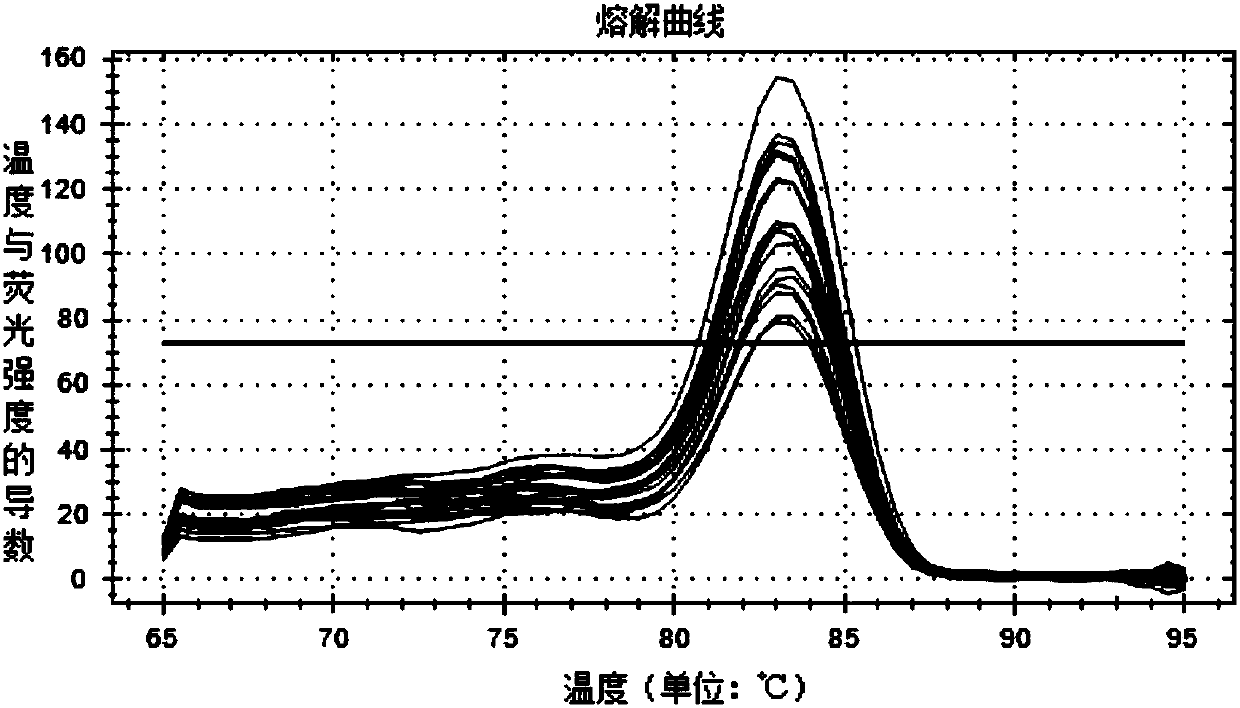
[0082] The known bacterial concentration is 0.48×10 9 Add 100uL of N. resiniferans in CFU / mL to 0.25g soil and mix evenly, so that the soil contains 1.92×10 N. resiniforans 8 CFU / g, the soil without N. resiniferans was set as the control. Utilize TIANGEN soil kit to extract soil DNA, carry out fluorescent quantitative PCR detection according to the condition of embodiment column 2, the result shows as follows Figure 5 As shown, by detecting the Ct value of the sample against the standard curve, the soil added with N. resiniferans can detect that the copy number of N. resiniforans in the soil sample is 1.59×10 8 copy / g, while N. resiniferous bacteria were not detected in the control soil.
[0083]
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com