DNA targeted capture array for leukemia chromosome aberration high-throughput sequencing
A DNA array and targeted capture technology, applied in the field of molecular biology, can solve the problems of high price, high cost, and low resolution, and achieve the effect of rapid screening and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] Example 1 Design of a DNA array for high-throughput targeted capture sequencing of leukemia chromosomal aberrations
[0015] Agilent SureSelect DNA Design Tool was used to design probes for 18 genes (see Table 2) involved in chromosomal aberrations. The parameters are set as follows:
[0016] Species: H.sapiens, hg19, GRCH37, February2009; Databases: RefSeq, Ensembl, CCDS, Gencode, VEGA, SNP,
[0017] CytoBand; Region: Entire Transcribed Region; Region Extension: 10 bases from 3’end and 10 bases from 5’end; Tiling density: 2×; Masking: Least Stringent; Boosting: Balanced.
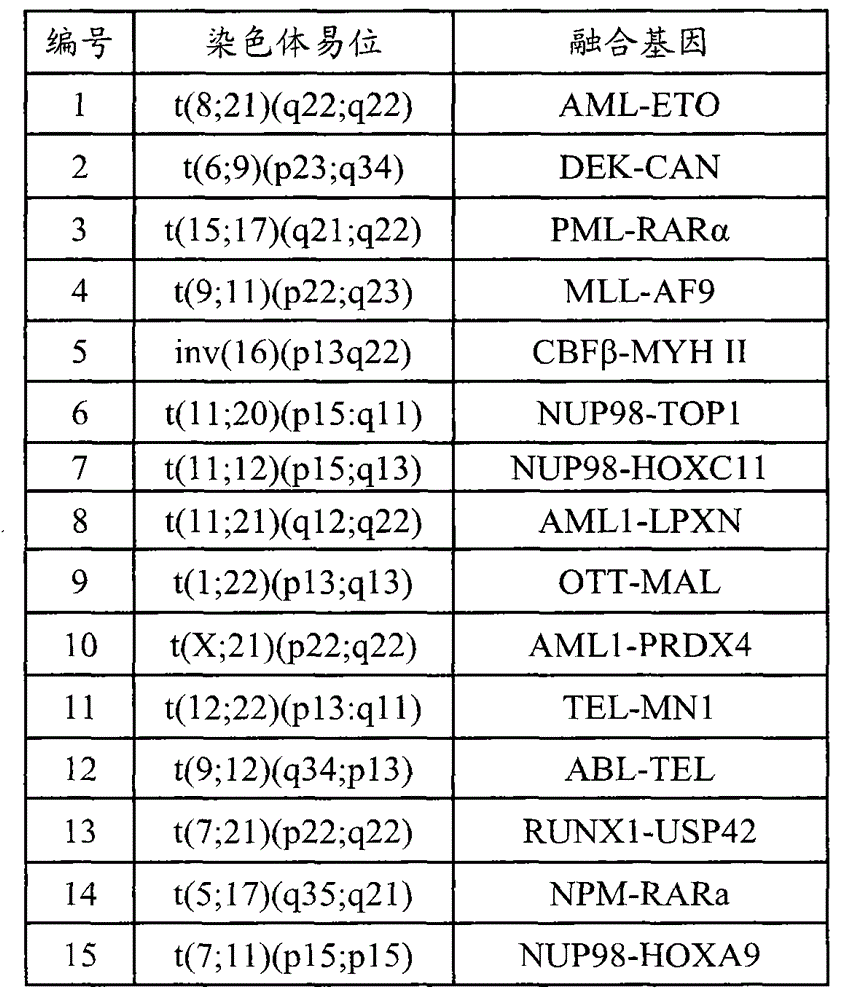
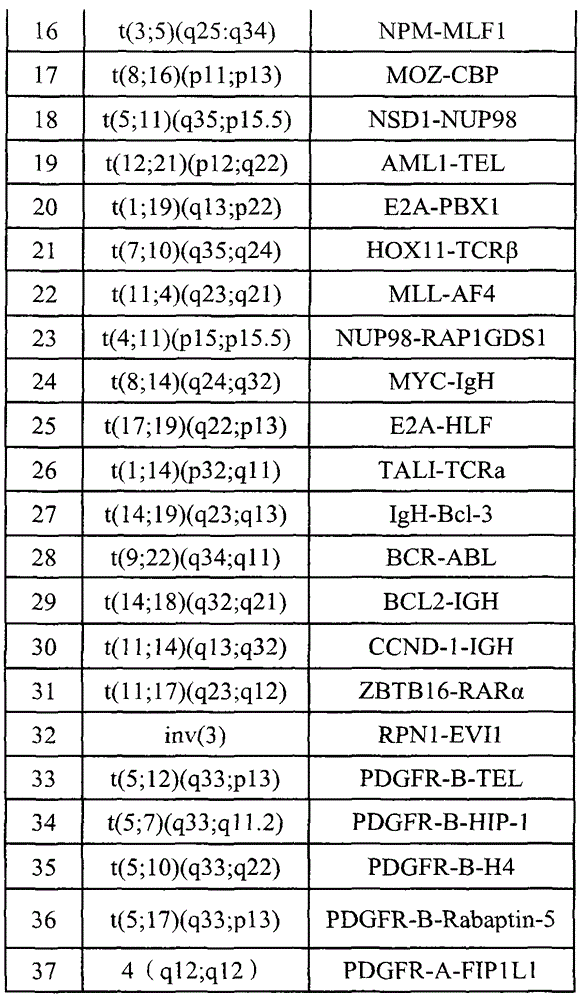
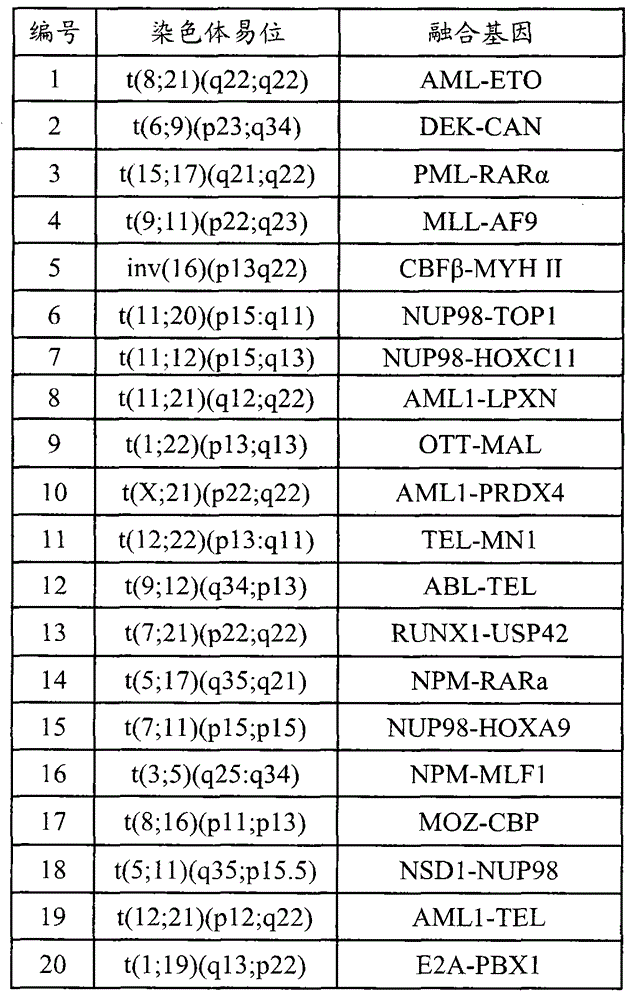
[0018] Table 2 Gene fragments captured by probes
[0019] serial number Gene chromosome start site stop site Region size (bp) 1 ABL1 (ABL) 9 133589258 133763072 173815 2 ETV6(TEL) 12 11802778 12048335 24558 3 IGHD (IGH) 14 106303089 106312020 8932 4 KAT6A(MOZ) 8 41786987 41909515 122529 5 KMT2A (MLL) 11 118307195 118397549 90355 ...
Embodiment 2
[0020] Embodiment 2 uses the present invention to detect clinical samples
[0021] 1. Extraction of bone marrow or peripheral blood samples to extract genomic DNA.
[0022] 2. Preparation before hybridization
[0023] 2.1 Genomic DNA is broken into small fragments of several hundred bases to build a library.
[0024] 2.2 Use Agencourt AMPure XP beads to purify the sample.
[0025] 2.3 Add base "A" to the 3' end of the DNA fragment.
[0026] 2.4 Use Agencourt AMPure XP beads to purify the sample.
[0027] 2.5 Add double-ended adapters.
[0028] 2.6 Use Agencourt AMPure XP beads to purify the sample.
[0029] 2.7 Amplify the library with adapters.
[0030] 2.8 Use Agencourt AMPure XP beads to purify the sample.
[0031] 2.9 Assess library quality.
[0032] 3. Hybridize and capture the product with the established library.
[0033] 4. The captured DNA fragments are denatured into single strands and combined with the adapter primers on the sequencing channel to form a bri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com