Method for detecting DAN methyltransferase activity based on strand displacement amplification and DNAzyme amplification
A technology of methyltransferase and strand displacement reaction, which is applied in the field of enzyme detection, can solve the problem of low sensitivity, and achieve the effect of wide application, good selectivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] M.SssI, HaeIII, HhaI and AluI methyltransferase, HpaII restriction endonuclease, KlenowFragment (3′-5′exo-) polymerase, Nb.BbvCI cleavage enzyme and S-adenosylmethionine from NEB Purchased. 5-azacytidine and 5-aza-2'-deoxycytidine were obtained from Sigma-Aldrich. Four kinds of deoxyribonucleic acid (dNTPs) and HEPES were purchased from Shanghai Shenggong Biotechnology Co., Ltd. The chemicals used in the experiment are all analytically pure, and no further purification is required during use. All solutions are prepared with ultrapure water (> 18.25MΩ·cm). The oligonucleotides in this work were synthesized and purified by Shanghai Shenggong Biotechnology Co., Ltd., and their sequences are listed in Table 1.
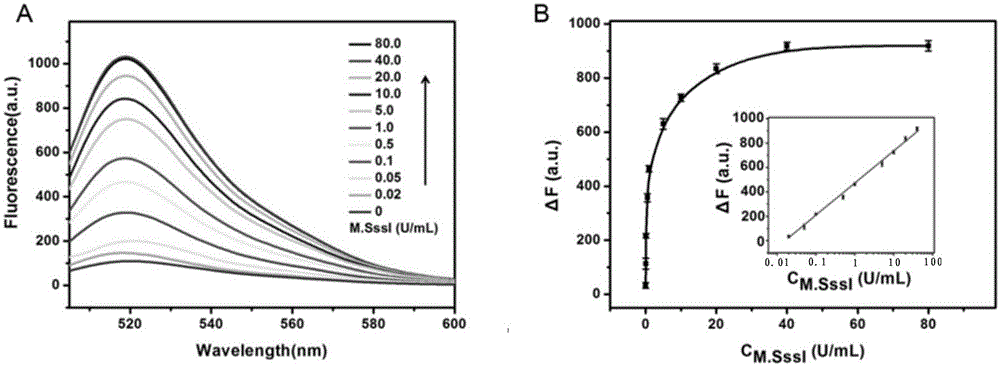
[0042] The fluorescence spectra of all samples were measured on the Hitachi F-7000 fluorescence spectrophotometer. The emission spectrum ranges from 505 to 600 nm, and the excitation wavelength is 494 nm. The fluorescence intensity at 518nm was used to evaluate th...
Embodiment 2
[0055] Principle analysis
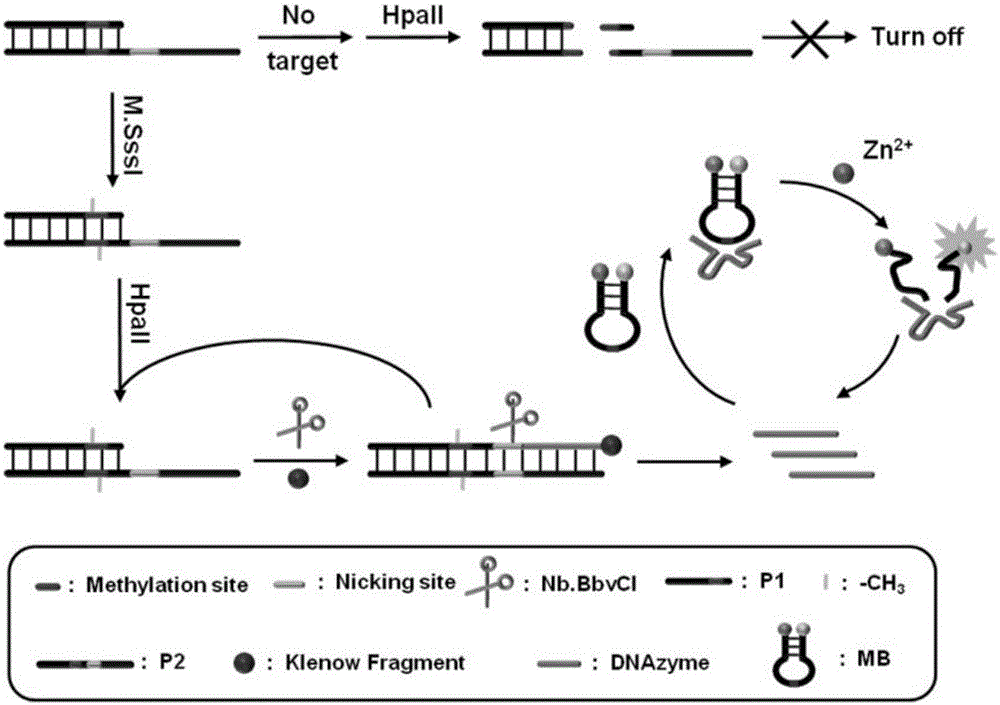
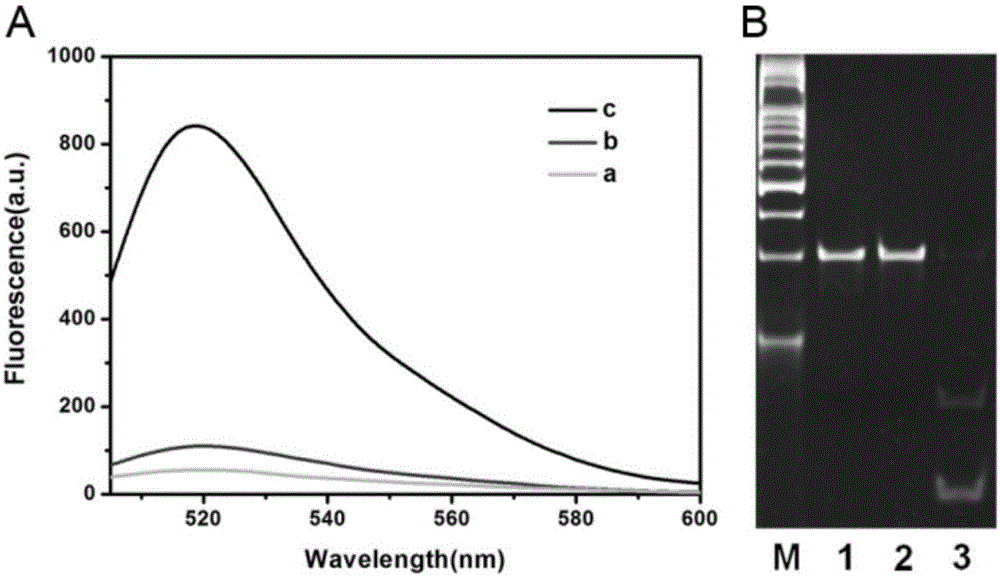
[0056] The principle of the proposed methyltransferase activity analysis is as follows figure 1 Shown. We have designed a trifunctional dsDNA probe, including methylation sites for DNA methyltransferase recognition, 8-17 DNAzyme complementary sequences for DNAzyme synthesis, and cleavage sites for cleavage by cleavage enzymes. First, M.SssI methyltransferase specifically recognizes and catalyzes the methylation of the trifunctional dsDNA probe to form methylated dsDNA. Subsequently, the HpaII restriction endonuclease specifically cleaves the remaining unmethylated dsDNA. Subsequently, under the action of KlenowFragment polymerase and dNTPs, P1 in the methylated dsDNA is used as a primer to initiate a polymerization reaction to form a complete DNA double strand with a cutting site. Next, Nb.BbvCI cleaves the entire DNA double-strand to create a new replication site for the next stage of polymerization. Through this repeated polymerization and cleavag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com