Detection primer set, detection kit and detection method for Vibrio parahaemolyticus, Photobacterium damsela and Nocardia seriolea
A technology of Nocardia japonica and a detection kit, which is applied in the field of detection of pathogenic bacteria in aquaculture animals, can solve problems such as cross-infection, omission, and hidden dangers of infection between shrimp and fish, and achieve rapid pathogen detection, low technical quality requirements, and application wide range of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The extraction of embodiment 1 bacterial genome DNA
[0034] Vibrio parahaemolyticus, Photobacterium mermaidi, and Nocardia aerevisiae (self-preserved in the Fishery Biological Disease Research Laboratory of South China Sea Fisheries Research Institute) were collected respectively, and the DNA extraction was carried out by the following steps:
[0035] (1) Resuspend the bacteria with 500-550 μL of TE buffer;
[0036] (2) Add 30 μL of SDS solution with a mass concentration of 10% and 15 μL of proteinase K (20 mg / mL) to the solution resuspended in step (1), mix well and incubate at 37° C. for 1 h;
[0037] (3) Add 100 μL of 5mol / L NaCl solution to the solution incubated in step (2), mix well and then add 80 μL of NaCl solution of LCTAB (dissolve CTAB in 0.5mol / L of NaCl solution, CTAB and all The mass volume ratio of the above NaCl solution is 1:20), and incubated at 65°C for 20min;
[0038] (4) Add phenol-chloroform-isoamyl alcohol (the volume ratio of phenol, chlorofo...
Embodiment 2
[0042] Example 2 Design and effectiveness detection of triple PCR detection primer sets for Vibrio parahaemolyticus, Photobacterium mermaid and Nocardia aerevisiae
[0043] 1. Select the specific genes (GI: 6714616, GI: 2988378, GI: 696553489) of Vibrio parahaemolyticus, Photobacterium mermaidi and Nocardia aerevisiae respectively, and use Primer Premier 5.0 software to analyze and design corresponding primer pairs. The primer pairs can specifically identify the specificity of the above-mentioned bacteria respectively, and the primer sequences are as follows:
[0044]
[0045]
[0046] 2. Effectiveness testing:
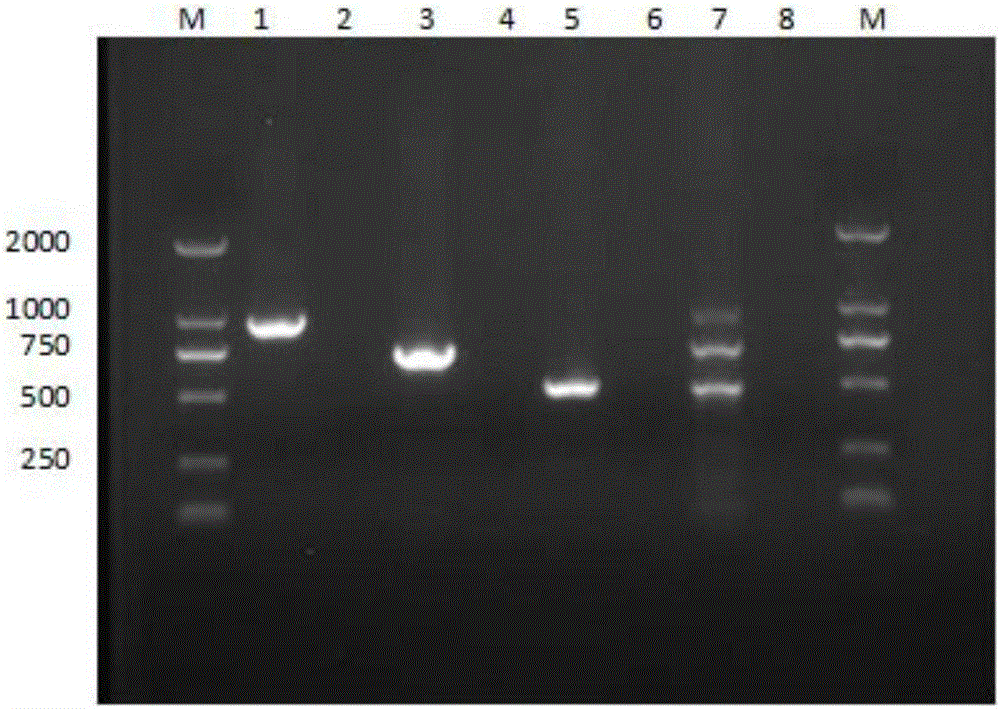
[0047] (1) Synthesize the primer set of the above-mentioned design, carry out single PCR verification to the DNA of Vibrio parahaemolyticus, Photobacterium mermaidus and Nocardia aerevisiae respectively with the primer of primer set, wherein single PCR reaction system is as follows:
[0048]
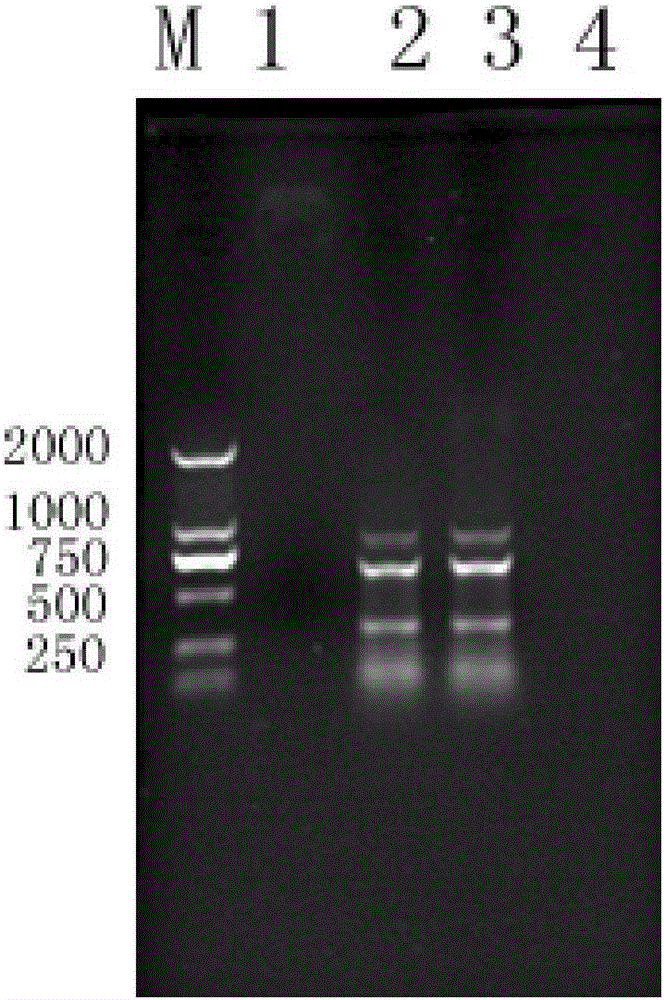
[0049] (2) Simultaneously perform triple PCR verification on Vibr...
Embodiment 4
[0062] Embodiment 4 detection kit
[0063] The detection kit of this embodiment can be used for rapid triple PCR diagnosis of Vibrio parahaemolyticus, Nocardiella aerevisiae and Photobacterium mermaidi in samples Bacillus detection primer set, proteinase K, SDS solution, phenol-chloroform-isoamyl alcohol mixed solution, isopropanol, ethanol with a volume concentration of 70%, TE buffer, CTAB NaCl solution, positive control substance and PCRDsMix ,in:
[0064] (1) Primer set for detection of Vibrio parahaemolyticus, Photobacterium mermaidi and Nocardia amberii: 1 tube contains Vibrio parahaemolyticus primers V.pa-F and V.pa-R, the nucleotide sequences of which are respectively shown in SEQ ID NO .1 and shown in SEQIDNO.2; 2 tubes are equipped with photobacterium mermaid primers N.se-F and N.se-R, and their nucleotide sequences are shown in SEQIDNO.3 and SEQIDNO.4 respectively; 3 tubes are equipped with amberjack The nucleotide sequences of the primers P.da-F and P.da-R for Ka...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com