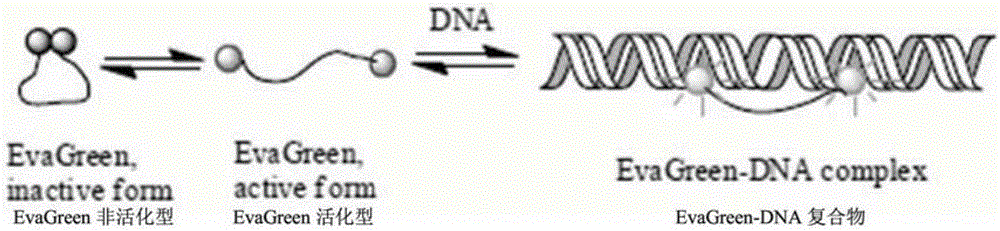
Quantitative PCR method adopting dye EvaGreen and dual HotStar polymerases
A hot-start enzyme and dye technology is applied in the field of quantitative PCR containing dye EvaGreen and dual hot-start enzymes to achieve the effect of inhibiting non-specific amplification, low price and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] In the real-time fluorescent quantitative PCR method, EvaGreen and the double hot start enzyme system have good repeatability, high sensitivity and precision.
[0056] (1) Construct a plasmid containing the β-globin gene and amplify the β-globin gene.
[0057] (2) Primer synthesis:
[0058] Forward primer: 5'-GAAGAGCCAAGGACAGGTAC-3'
[0059] Reverse primer: 5'-CAACTTCATCCACGTTCACC-3'
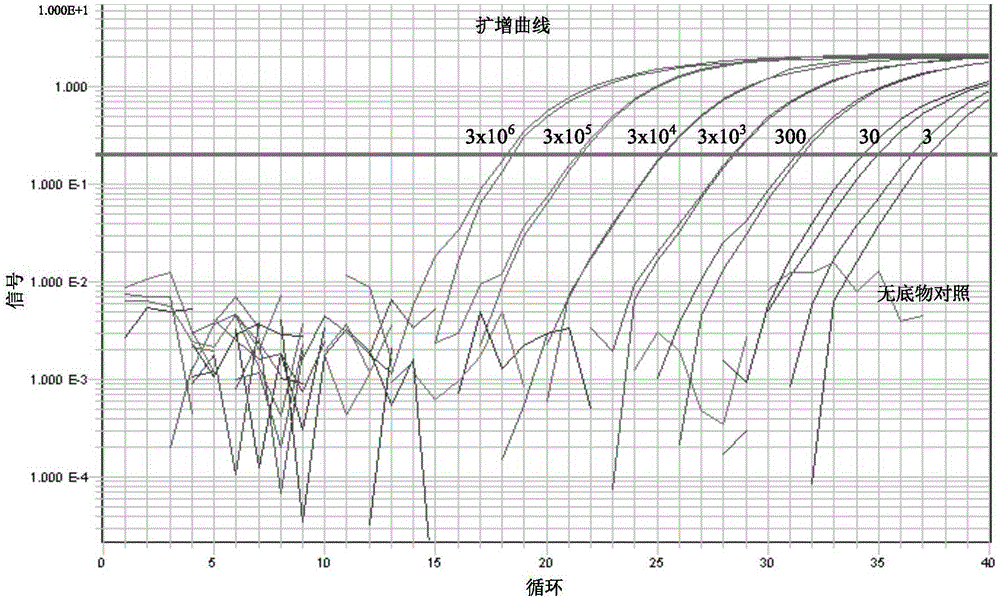
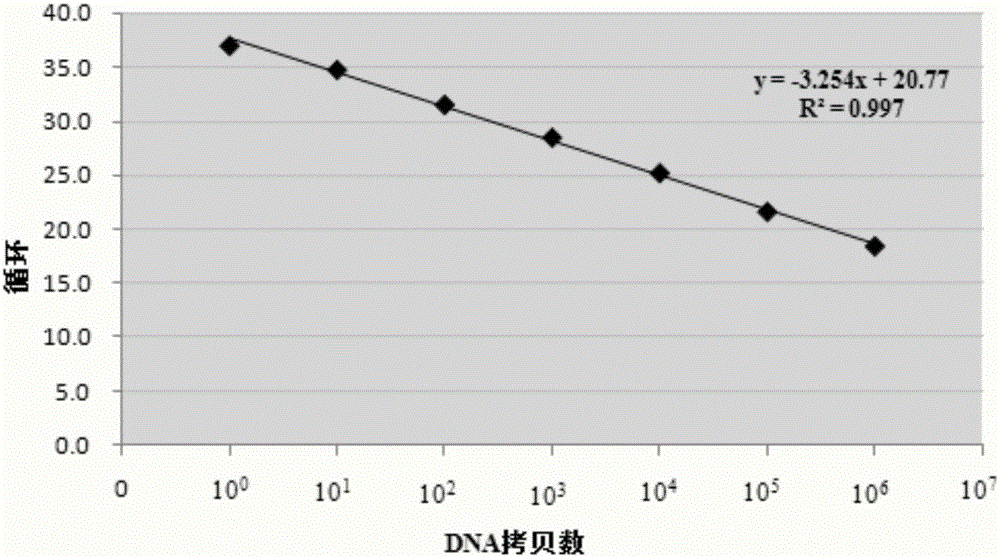
[0060] (3) Prepare the plasmid template as 3x10 6 , 3x10 5 , 3x10 4 , 3x10 3 , 3x10 2 , 3x10 1 , 3x10 0 The amount of template for each molecule, NTC is no substrate control, real-time fluorescence quantitative PCR reaction is carried out by using forward primer and reverse primer together with the PCR reaction master solution containing EvaGreen and dual hot start enzyme.
[0061] (4) Establishment of the PCR reaction system, the reaction system is 20 μL in total, and each reagent is added referring to the table below.
[0062] Table 1 PCR reaction system
[0063]
[0064] ...
Embodiment 2
[0072] In the real-time fluorescent quantitative PCR method, the dual hot start enzyme system has stronger sensitivity and higher amplification efficiency.
[0073] (1) Construct a plasmid containing the β-globin gene and amplify the β-globin gene.
[0074] (2) Primer synthesis is the same as in Example 1.
[0075] (3) Prepare the plasmid template as 3x10 4 , 3x10 3 , 3x10 2 , 3x10 1 , 3x10 0 The template amount of each molecule was amplified by dual hot-start enzyme and single hot-start enzyme respectively, and real-time fluorescent quantitative PCR reaction was carried out in a system with other components unchanged.
[0076] (4) PCR reaction system: dual hot-start enzymes, chemically modified DNA polymerase and antibody-modified DNA polymerase were used as hot-start enzymes, and other components were the same as in Example 1.
[0077] (5) qPCR reaction is the same as in Example 1.
[0078] (6) Experimental results
[0079] The result is as Figure 4 shown. Figure...
Embodiment 3
[0081] In the real-time fluorescent quantitative PCR method, EvaGreen has higher specificity and better repeatability.
[0082] (1) Prepare DNA to be tested; extract DNA from human blood according to conventional methods, and amplify uridine diphosphate glucuronosyltransferase gene (UGT1A1).
[0083] (2) Primer synthesis:
[0084] Forward primer: 5'-ACCTCTAGTTACATAACCTGA-3'
[0085] Reverse primer: 5'-AATAAACCCGACCTCACCAC-3'
[0086] (4) PCR reaction system: EvaGreen, SYBRGreenI and GelGreenI were used as fluorescent dyes, and other components were the same as in Example 1.
[0087] (5) The qPCR reaction procedure is the same as in Example 1.
[0088] (6) Experimental results
[0089] In this example, the difficult-to-amplify UGT1A1 gene tested by the inventors was selected as a template, and three double-stranded DNA-binding dyes, EvaGreen, SYBRGreenI and GelGreenI, were used to compare their specificity and repeatability. As shown in Figure 5. Figure 5A-1 and 5A-2 The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com