Method capable of extracting high-quality genome DNA from fungus by adopting CTAB process
A high-quality, genomic technology, applied in recombinant DNA technology, DNA preparation, etc., can solve problems such as difficulty in meeting molecular experiments, insufficient DNA concentration and purity, and limited success rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
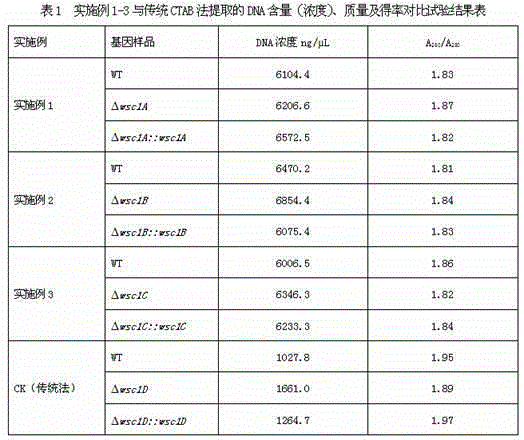
Examples
Embodiment 1
[0028] Prepare 1×10 with sterilized 0.02% Tween water 7 spore suspension per ml, take 100 μL of the suspension and inoculate it on the cellophane on the surface of the SDAY plate, and incubate at 25°C for 2.5 days. Then scrape the mycelium on the cellophane and put it into a mortar pre-cooled in a -70°C refrigerator, add 0.3 g of sterilized fine sand of 60 mesh and add liquid nitrogen for rapid grinding for 3 minutes until the mycelium finally becomes a silky ultrafine powder (600-1000 mesh), and then follow the steps of CTAB method to minimize exposure to the air during the extraction process, and use a refrigerated centrifuge to centrifuge at 4°C for all centrifugation processes.
Embodiment 2
[0030] Prepare 1×10 with sterilized 0.02% Tween water 7spore suspension per ml, take 150 μL of the suspension and inoculate it on the cellophane on the surface of the SDAY plate, add 0.1 g of 70-mesh sterilized fine sand, and incubate at 25°C for 2.5 days. Then scrape the mycelium on the cellophane into a mortar pre-cooled in a -70°C refrigerator, add liquid nitrogen for rapid grinding for 4 minutes until the mycelium finally presents a silky ultrafine powder (600-1000 mesh), and then follow the The CTAB method is operated step by step, and the exposure to the air is minimized during the extraction process. All centrifugation processes are centrifuged at 4°C in a refrigerated centrifuge.
Embodiment 3
[0032] Spread a layer of sterilized cellophane on the SDAY board, add 120 μL of 0.02% Tween water, and then add an appropriate amount (0.2 g) of 80 mesh fine sand containing the target strain to the SDAY cellophane and coat the board evenly with a coating stick; 25 Cultivate for 3 days. Then scrape the mycelium on the cellophane into a mortar pre-cooled in a -70°C refrigerator, add liquid nitrogen for rapid grinding for 5 minutes until the mycelium finally presents a silky ultrafine powder (600-1000 mesh), and then follow the The CTAB method is operated step by step, and the exposure to the air is minimized during the extraction process. All centrifugation processes are centrifuged at 4°C in a refrigerated centrifuge.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com