Pseudomonas aeruginosa detection kit and application thereof
A technology of Pseudomonas aeruginosa and a detection kit, which is applied in the determination/inspection of microorganisms, microorganisms, biochemical equipment and methods, etc., can solve the problems of unsatisfactory reliability and repeatability, and achieve short detection time and high sensitivity High and specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
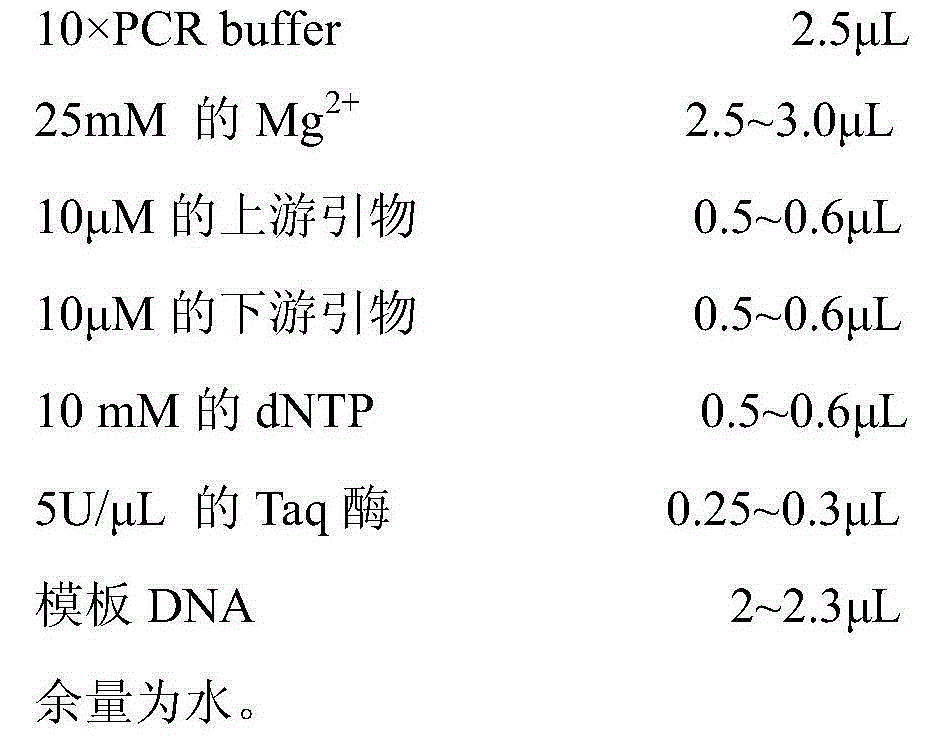
[0028] A detection kit for Pseudomonas aeruginosa, comprising an upstream primer of the sequence shown in SEQID01, a downstream primer of the sequence shown in SEQID02, a selective enrichment solution, a PCR reaction reagent, a positive control, a negative control and a blank control, the The proportions of the components in the PCR reaction system of the kit are as follows: 25 μL of the PCR reaction system includes:
[0029]
[0030]
[0031] The amplification temperature program of the above PCR reaction is as follows: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 1 min, annealing at 60°C for 30 s, extension at 72°C for 15 s, 35 cycles; extension at 72°C for 7 min.
[0032] The formula ratio of the above selective enrichment solution is: 1L of the selective enrichment solution contains tryptone 10g, sodium chloride 10g, yeast extract 5g, cetyltrimethylammonium bromide 0.2g and naphthyridone Acid 0.015g.
[0033] The method for detecting using the above-...
Embodiment 2
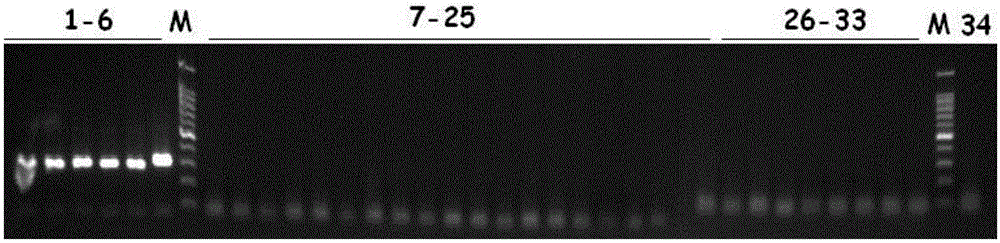
[0037] Example 2: Verification of the specificity of the detection primers in the present invention.
[0038]This embodiment extracts 6 strains of Pseudomonas aeruginosa (Pseudomonasaeruginosa) ATCC27853, ATCC15442, ATCC9027, CGMCC1.10274, CGMCC1.10452, XMZJ~1 (laboratory isolation, identified as Pseudomonas aeruginosa by VITEK2Compact), 19 Other bacteria of the genus Pseudomonas [P.azotoformans CGMCC1.1792, P.mucidolens CGMCC1.1795, Pseudomonas alcaligenes CGMCC1.3361, Pseudomonas putida (P.putida) CGMCC1.3301, Pseudomonas pseudoalcaligenes (P.pseudoalcaligenes) CGMCC1.2935, Pseudomonas stutzeri (P.stutzeri) CGMCC1.3340, door Pseudomonas dorsalis (P.mendocina) CGMCC1.2965, Pseudomonas asplenii (P.asplenii) CGMCC1.4995, Pseudomonas green needle (P.chlorophis) CGMCC1.2887, Pseudomonas chicory Bacteria (P.cichorii) CGMCC1.4934, Pseudomonas flavescens (P.flavescens) CGMCC1.6138, Pseudomonas citronellolis (P.citronellolis) CGMCC1.6143, Pseudomonas jaszii (P. jessenii) CGMCC1.885...
Embodiment 3
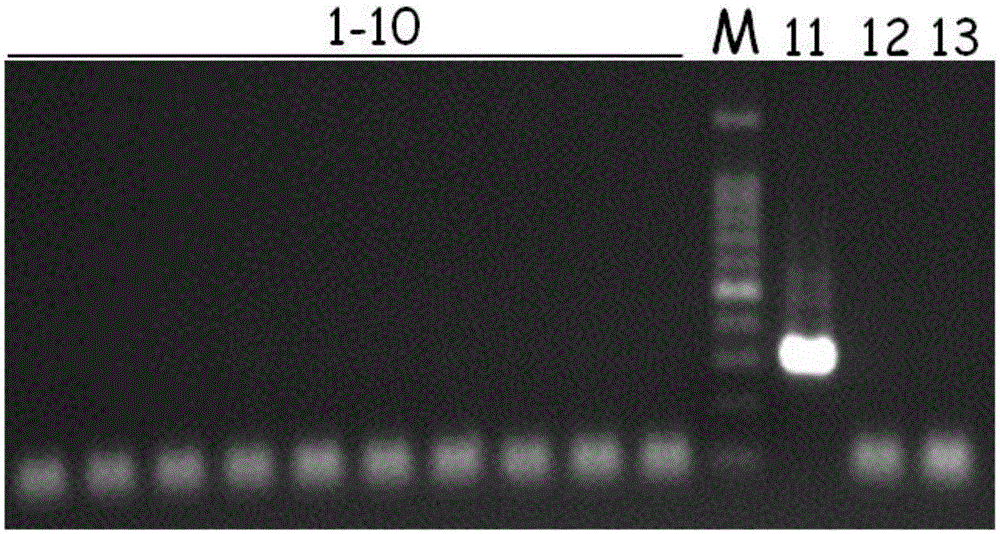
[0055] Example 3: Detection of Pseudomonas aeruginosa in bottled drinking purified water.
[0056] This example uses the specific primers and PCR method of Example 1 of the present invention to detect 10 parts of commercially available bottles and barrels of drinking purified water, and compares the parallel detection results of traditional detection methods (GB19298-2014) to verify the establishment of the present invention reliability of the detection method.
[0057] The primers and PCR method used are the same as in Example 1.
[0058] The main testing instruments used:
[0059] Milliflex~plus microbial filtration system (Millipore, Germany), 250 mL filter cup with 0.45 μm pore size filter membrane, micropipette (10 μL, 100 μL, 1000 μL, Eppendorf), centrifuge (5424R, Eppendorf, Germany), gradient PCR amplification Multiplier (Veriti, American ABI Company), electrophoresis instrument (TanonEPS300, Shanghai Tianneng Technology Co., Ltd.), gel imager (Tanon3500, Shanghai Ti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com