DNA methylation detection method of specific gene segment
A specific gene and detection method technology, applied in the field of DNA methylation level detection, can solve problems such as difficult to apply to clinical routine detection applications, CpG site methylation, complex data processing, etc., to reduce detection costs and have good applicability , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
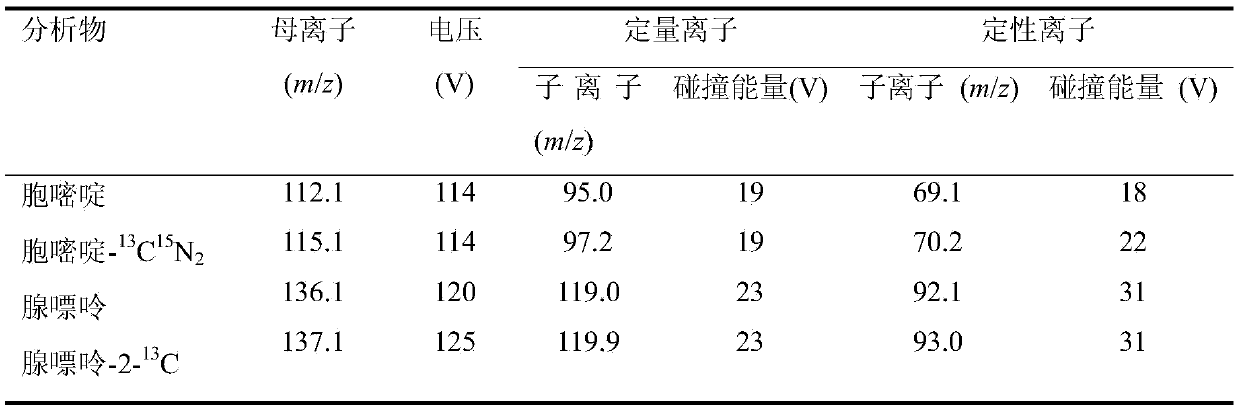
[0032] Example 1: This method was used to measure the methylation level in exon 2 of the p16 gene in tumor tissue and paracancerous tissue. Note: In this example, the chemical cleavage method is used to cleavage the gene fragment into a single base.
[0033] 1. DNA extraction and bisulfite treatment
[0034] The total genomic DNA of the biological samples was extracted using a commercial DNA extraction kit, and the purity and concentration of the DNA solution were detected by agarose gel electrophoresis and UV-visible spectrophotometry.
[0035]Take a certain amount of genomic DNA, and use a commercial methylation conversion kit for bisulfite treatment to convert unmethylated cytosine (C) into uracil (U), while methylated cytosine constant.
[0036] 2. Acquisition of exon 2 fragment of p16 gene:
[0037] Genomic DNA treated with bisulfite was used as a template, and a pair of primers without CpG sites (upstream: 5'-TGGTAGGTTATGATGATGGGTAG-3', downstream: 5'-CTCAAATCATCAATCC...
Embodiment 2
[0057] Example 2: This method is used to measure the methylation level in exon 2 of p16 gene in tumor tissue. Note: In this example, the enzymatic hydrolysis method was used to cleavage the gene fragments into single nucleosides.
[0058] While keeping the steps of DNA extraction, sulfite treatment, and PCR amplification of gene fragments unchanged, we performed enzymatic hydrolysis on the exon 2 fragment product of p16 gene to obtain single nucleosides, and then used the preferred liquid phase The tandem mass spectrometry program detects the content of the above-mentioned nucleosides, and determines the DNA methylation level of the gene fragment through a formula.
[0059] (1) Cleavage of DNA gene fragments: Denature the gene fragments at 95°C for 5 minutes, and cool on ice for 2 minutes; then add S1 endonuclease and its buffer, and incubate at 37°C for 16 hours; then add alkaline phosphoric acid Salt and its buffer, continue to incubate at 37°C for 4h. The nucleosides gene...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com