Regulatable gene expression
A gene expression and adjustable technology, which is applied in the determination/testing of microorganisms, growth factors/inducing factors, nucleotide libraries, etc., and can solve the problem that large groups are not effective
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
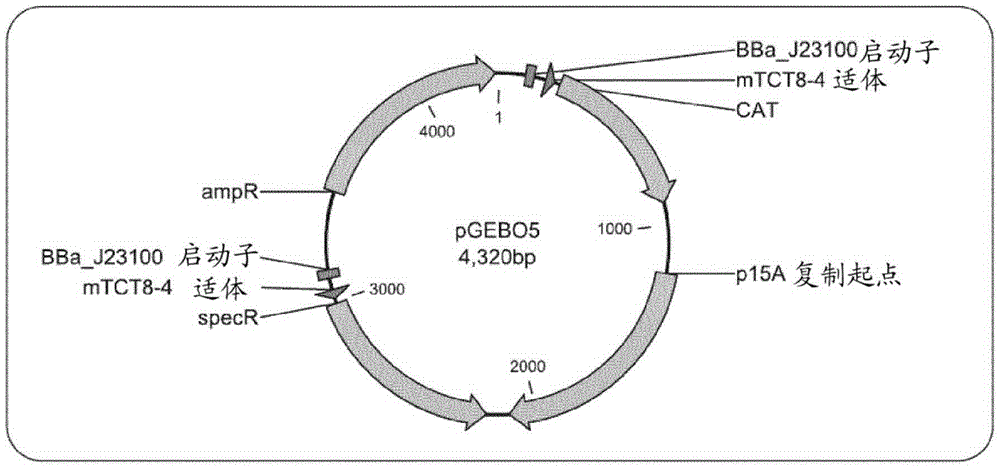
[0154] Example 1. Construction of double riboswitch selection vector pGEBO5.1
[0155] pGEBO5.1 is a third generation selection vector based on the second generation selection vector pGEBO4.1, and pGEBO4.1 is based on the first generation selection vector pGEBO4. pGEBO4 comprises the vector pGEBO2 containing the 'riboswitch platform'.
[0156] Construction of pGEBO2: 221 base pair DNA sequence constituting the 'riboswitch platform' ( figure 1 ) was artificially synthesized by Integrated DNA Technologies and delivered on the pIDTSMART vector, resulting in pGEBO2. In addition to the riboswitch platform, the 5' end of the riboswitch includes a Flippase Recognition Target (FRT) sequence (5'-GAAGTTCCTATTTCTCTAGAAAGTATAGGAACTTC-3' (SEQ ID NO: 19)).
[0157] Construction of pGEBO4: pGEBO4 was established by circular USER fusion of three PCR products: (1) FRT-riboswitch platform amplified from pGEB02 with primers GEBO4 / 5 (forward / reverse); (2) with primer GEBO6 / 2 region containing...
Embodiment 2
[0160] Example 2. Determination of spontaneous resistance in single and double selection systems
[0161] pGEBO5.1 was transformed into E. coli Dh10B cells (forming EcpGEBO5.1 ) using standard methods known to the skilled person. To test whether the dual selection system resolved the problem of spontaneous resistance, up to 1.3x10 8 Plating of EcpGEBO5.1 cells ('double selection'). As a control, plating on plates containing only chloramphenicol (30 μg / mL) or spectinomycin (80 μg / mL) was each performed in triplicate (‘single selection’) (see Table 1). Each plate was incubated for 72 hours. In the 10 plates subjected to double selection, no colonies were observed within the first 48 hours of incubation. During the 24 h incubation, each of the three plates containing only chloramphenicol produced numerous colonies, as did the plate containing only spectinomycin. When the 1.3×10 8Spontaneous resistance was observed in the double selection system when each cell was plated and ...
Embodiment 3
[0164] Example 3. Growth response of EcpGEBO5.1 to different xanthine alkaloids
[0165] The growth response of EcpGEBO5.1 to 4 different xanthine alkaloids was determined empirically using a selection assay with different concentrations of the xanthine-based compounds theophylline, xanthine, theobromine and caffeine.
[0166] Procedure: Plate 300-400 colony-forming units in a mixture containing ampicillin (50 µg / mL), spectinomycin (80 µg / mL), chloramphenicol (30 µg / mL), and xanthine-based compounds at indicated concentrations (see Table 2) On the flat plate: predetermine the dissociation constant (K d ) (Jenison et al., 1994). No selection refers to plates without spectinomycin and chloramphenicol. (+) means growth was observed. (-) means that no growth was observed. (approximately) means that reduced growth was observed.
[0167] Table 2 - Characterization of the specificity of the riboswitch aptamer domain in pGEBO5.1.
[0168]
[0169] These results indicate that ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com