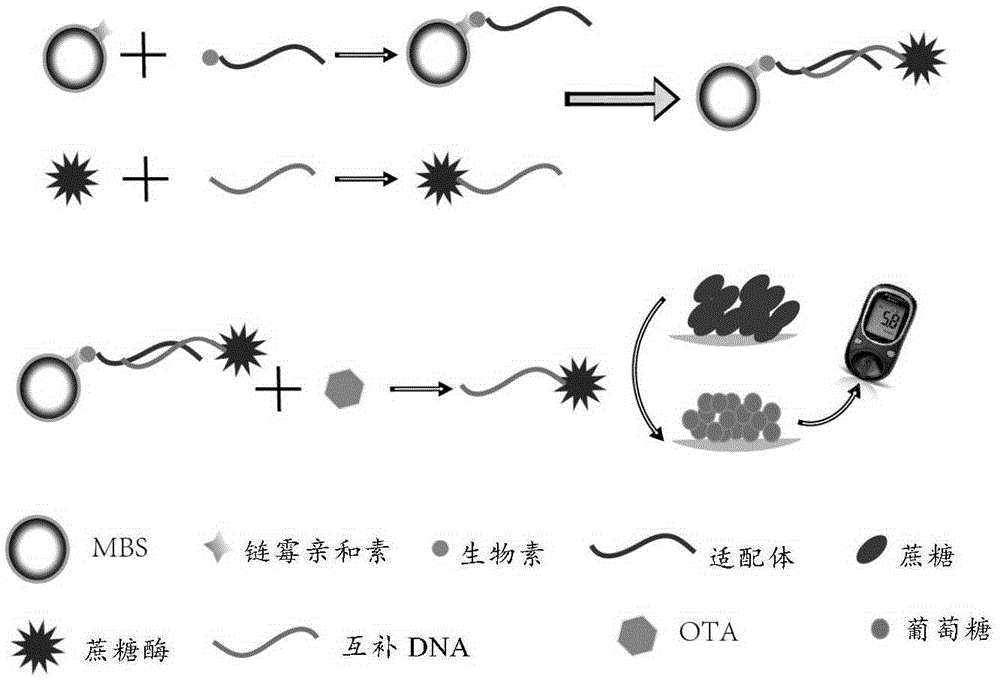
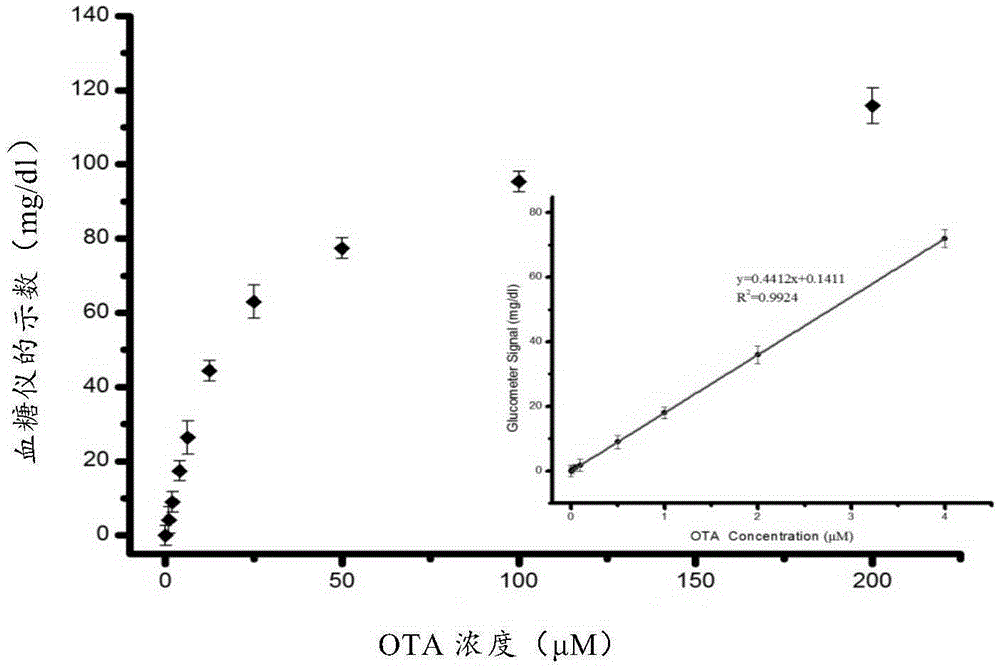
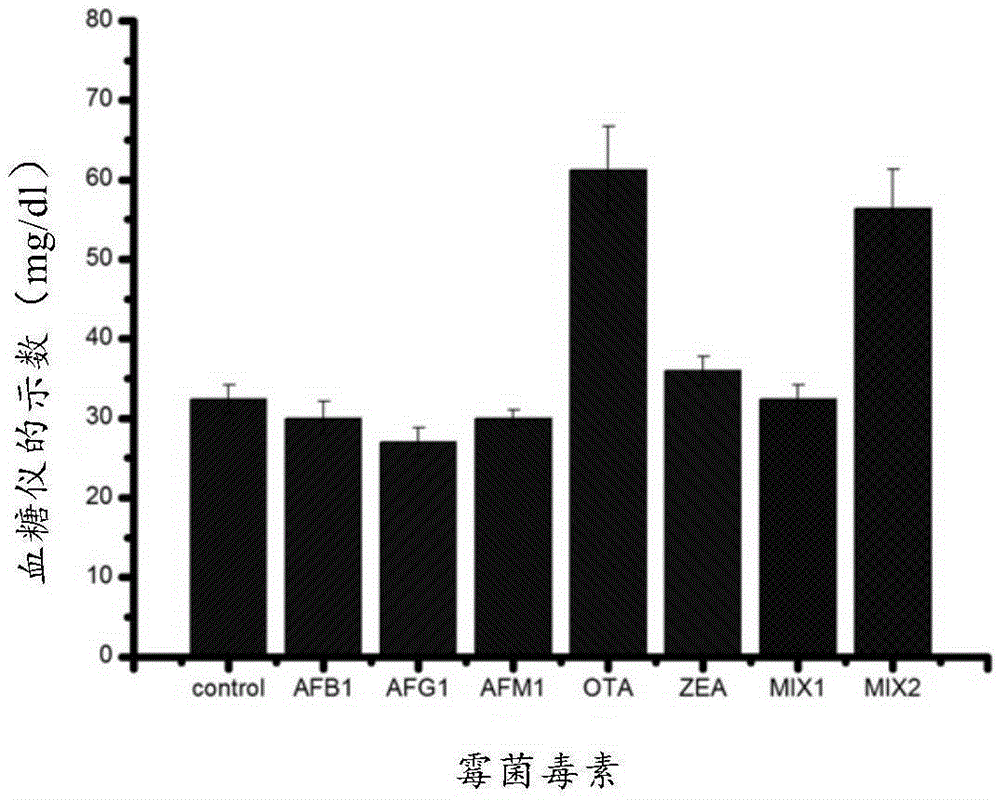
Method for quantitatively detecting ochratoxin A
A technology for quantitative detection of ochratoxin, applied in the field of quantitative detection of ochratoxin A, aptamer biosensor combined with blood glucose meter for quantitative detection of ochratoxin A, achieving good specificity, low detection limit, and good application prospects Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Synthesis of aptamer biosensor and detection of ochratoxin A in combination with a blood glucose meter
[0046] 1. Experimental method
[0047] 1.1 Synthesis of DNA-sucrase polymer
[0048] 1.1.1 Activation of sucrase molecules
[0049] Take 400 μl of 20 mg / ml sucrase (buffer B) and mix with 1 mg sulfo-SMCC, vortex and shake for 5 min, place on a constant temperature mixer, and react at room temperature for 2 h.
[0050] 1.1.2 Activation of DNA molecules
[0051] Take 100 μl of 100 μM complementary DNA (thiol-DNA, 5'-CCCACACCCGATCAAAAAAAAAAAA-SH-3'), 2 μl of 0.1M bufferB, 2 μl of 30mM TCEP (ultrapure water) and add it to a 1.5ml centrifuge tube, vortex and mix well, and place it on a constant temperature mixer. React at room temperature for 1 hour (1) Treatment of complementary DNA: Centrifuge the synthesized solid DNA (4°C, 12000 rcf, 5 min), add 345 μl of ultrapure water as required, and vortex slightly to obtain 345 μl of 100 μM DNA solution; 2) Preparat...
Embodiment 2
[0079] Synthesis of Example 2 Aptamer Biosensor
[0080] 1.1 Synthesis of DNA-sucrase polymer
[0081] 1.1.1 Activation of sucrase molecules
[0082] Take 300 μl of 20 mg / ml sucrase (buffer B) and mix with 0.5 mg sulfo-SMCC, vortex and shake for 5 min, place on a constant temperature mixer, and react at room temperature for 1 h.
[0083] 1.1.2 Activation of DNA molecules
[0084] Take 80 μl of 100 μM complementary DNA (thiol-DNA, 5'-CCCACACCCGATCAAAAAAAAAAAA-SH-3'), 1 μl of 0.1M bufferB, 1 μl of 30mM TCEP (ultrapure water) and add it to a 1.5ml centrifuge tube, vortex and mix well, and place it on a thermostatic mixer. Reaction at room temperature for 0.5h.
[0085] 1.1.3 Synthesis of DNA-sucrase polymer
[0086] Centrifuge the reaction solution of sucrase-SMCC and thiol-DNA (25°C, 12000rcf, 5min), draw the supernatant, and add them to ultrafiltration tubes (Amicon-100K for sucrase-SMCC; Amicon-100K for thiol-DNA) 3K), centrifuge (25°C, 12000rcf, 10min), wash 8 times with...
Embodiment 3
[0094] Example 3 Synthesis of Aptamer Biosensors
[0095] 1.1 Synthesis of DNA-sucrase polymer
[0096] 1.1.1 Activation of sucrase molecules
[0097] Mix 500 μl of 20 mg / ml sucrase (buffer B) with 2 mg of sulfo-SMCC, vortex for 5 min, place on a constant temperature mixer, and react at room temperature for 3 h.
[0098] 1.1.2 Activation of DNA molecules
[0099] Take 120 μl of 100 μM complementary DNA (thiol-DNA, 5'-CCCACACCCGATCAAAAAAAAAAAA-SH-3'), 3 μl of 0.1M bufferB, 3 μl of 30mM TCEP (ultrapure water) and add it to a 1.5ml centrifuge tube, vortex and mix well, and place it on a constant temperature mixer. Reaction at room temperature for 2h.
[0100] 1.1.3 Synthesis of DNA-sucrase polymer
[0101] Centrifuge the reaction solution of sucrase-SMCC and thiol-DNA (25°C, 12000rcf, 5min), draw the supernatant, and add them to ultrafiltration tubes (Amicon-100K for sucrase-SMCC; Amicon-100K for thiol-DNA) 3K), centrifuge (25°C, 12000rcf, 10min), wash 8 times with bufferA; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com