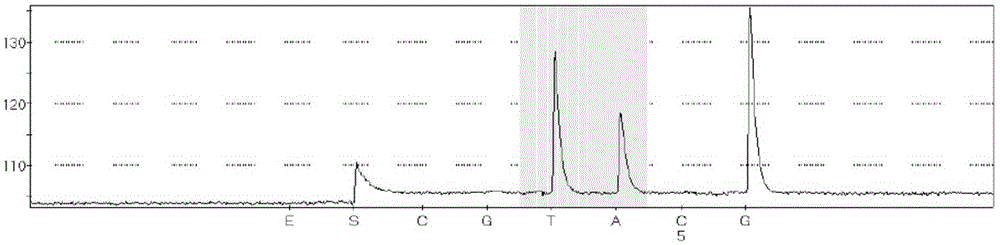
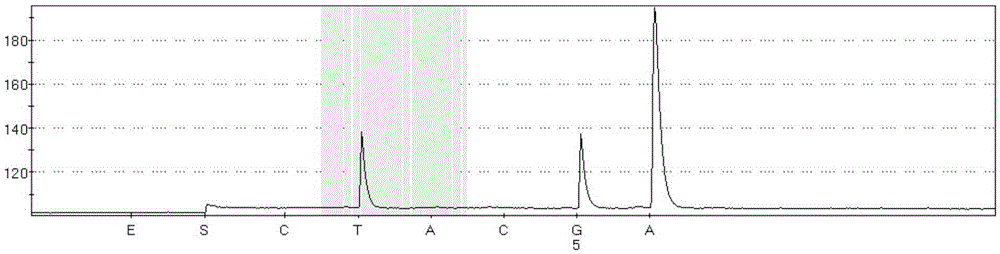
Kit for detecting BRAF gene V600E trace mutation through pyrosequencing technique and application of kit
A pyrosequencing and technical detection technology, which is applied in the field of pyrosequencing technology to detect BRAF gene V600E trace mutation kits, can solve the problems of low positive detection rate and cumbersome operation, and achieves easy interpretation, simple operation, and reduced sample collection. desired effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Reagents.
[0038] (1) DNA extraction reagents:
[0039] Purchased from QIAGEN Company.
[0040] (2) Reaction solution:
[0041] PCRBuffer: purchased from Fermentas, USA;
[0042] Primers SEQIDNO: 1-12, synthesized by Shanghai Yingjun Biotechnology Co., Ltd.;
[0043] MgCl 2 : purchased from the U.S. Fermentas company;
[0044] 0.2mMdNTPs: purchased from Fermentas, USA;
[0045] 2U / μL TaqDNA polymerase: purchased from Fermentas, USA.
[0046] (3) Reagent for single-strand purification:
[0047] 75% (v / v) ethanol solution: purchased from Hangzhou Changzheng Chemical Reagent Co., Ltd.;
[0048] 0.2M NaOH: purchased from Shanghai Shisi Hewei Chemical Co., Ltd.;
[0049] 10mM Tris-Acetate (pH7.6): Tris-base was purchased from Sigma, USA, and anhydrous acetic acid was purchased from Hangzhou Chemical Reagent Co., Ltd.;
[0050] Binding buffer: 10mM Tris-HCl (Tris-base was purchased from Sigma, USA, hydrochloric acid was purchased from Hangzhou Chemical R...
Embodiment 2
[0057] Embodiment 2: detection method.
[0058] Instruments: Bio-RadS1000PCR instrument, BeckmanMicrofuge22R desktop microrefrigerated centrifuge, Beijing Liuyi agarose gel electrophoresis instrument, Shanghai Peiqing gel imaging system, QIAGENPyroMarkQ96ID sequencer.
[0059] (1) Extract DNA from paraffin biopsy tissue samples, the steps are as follows:
[0060] a. Take tissue DNA, add tissue lysate, proteinase K, and incubate at 56°C for 2 hours;
[0061] b. Transfer the lysate to a DNA adsorption column and centrifuge at 12,000 rpm for 1 minute;
[0062] c. Add 500uL rinse solution 1 and centrifuge at 12000 rpm for 30 seconds;
[0063] d. Add 500uL rinse solution 2 and centrifuge at 12000 rpm for 30 seconds;
[0064] e. Transfer the DNA adsorption column to a new 1.5mL centrifuge tube, add 50uL deionized water, and centrifuge at 12000 rpm for 1 minute to obtain genomic DNA.
[0065] (2) using the genomic DNA obtained in step (1) as a template, and using EGFR-specific prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com