Application of differential expression of gene in oral cancer diagnosis
A gene expression, oral cancer technology, applied in the application field of differential expression in the diagnosis of oral squamous cell carcinoma, can solve the problems of radical surgery, no significant increase in the five-year survival rate, etc., and achieves strong specificity and good application prospects. , good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
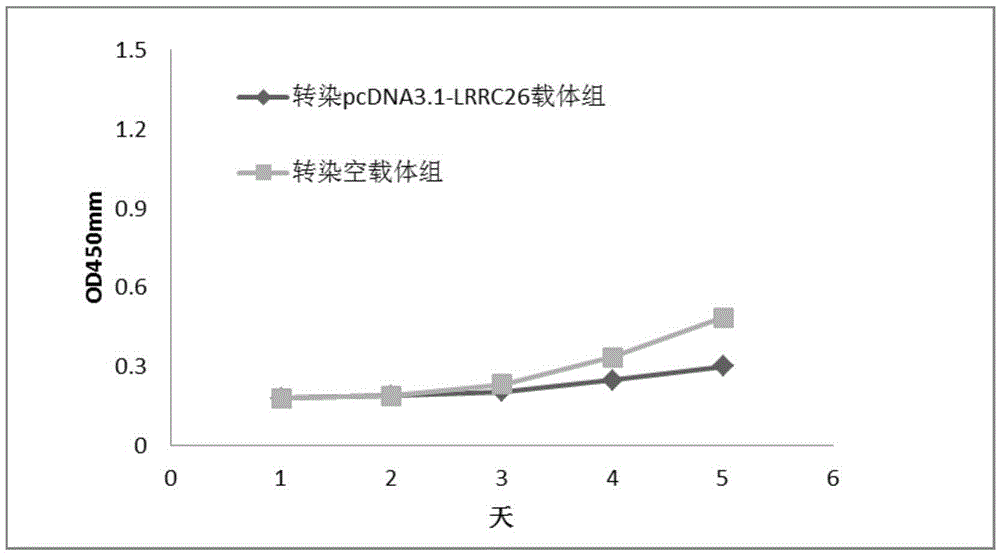
Embodiment 1
[0032] Oral cancer and normal tissues were collected, grouped and numbered: 3 cases of normal tissue blocks (numbered N1, N2, N3) and 2 cases of cancer tissue blocks and paracancerous tissue blocks (C1, C2 and P1, P2).
[0033] The three groups of samples were subjected to RNA extraction, followed by agarose gel electrophoresis after RNA extraction. From the results of electrophoresis, the quality of the extracted RNA samples was preliminarily considered to be qualified and could be used for further transcriptome analysis. Further, the extraction of RNA samples was detected by NanoDrop1000 spectrophotometer, and the results are shown in Table 1. Each sample volume was 30 μl. Sample requirements for RNA-seq sequencing: OD260 / OD280 is 1.8-2.2. RIN (RNA integrity number) refers to the RNA integrity index, which is an important indicator of RNA quality. It can be seen from Table 1 that all the samples in this study met the requirements of transcriptome sequencing.
[0034] Tab...
Embodiment 2
[0036] Example 2 High-throughput transcriptome sequencing and analysis
[0037] Sequencing platform: Illumina's HiSeq2500 high-throughput sequencing platform
[0038] Transcriptome sequencing and matching:
[0039] The 3 samples of cancerous tissue, paracancerous tissue and normal tissue we analyzed came from 2 cases of oral cancer patients and 3 cases of healthy people. These two sets of samples were subjected to high-throughput cDNA sequencing. From these 3 tissues we get 4.85×10 7 ,4.91×10 7 , 4.46×10 7 read pairs. We used TopHat to match reads to the UCSC reference human genome (hg19), and the unique matched read pairs ranged from 4.04 × 10 7 to 4.49×10 7 The proportion of reads matching to Ensembl reference genes ranged from 81% to 89%. The average coverage of our sequencing depth is about 130 times that of the human genome (according to the total length of the exon region uniquely annotated in the Ensembl database, it is about 1.13×10 8 ), and only 1% of the rea...
Embodiment 3
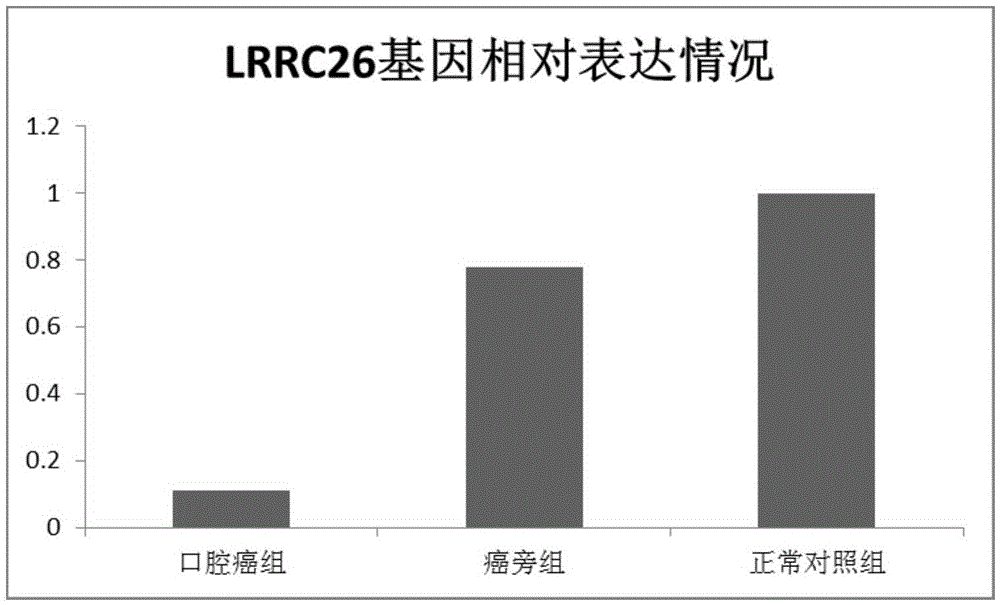
[0044] Example 3 Cancer tissue, paracancerous tissue and normal tissue LRRC26 gene expression
[0045] 1. Materials and methods
[0046] 1. Materials
[0047] Oral cancer patients were hospitalized from 2010 to 2014, and the cancer tissues and paracancerous tissues of 49 patients with oral cancer and 23 normal tissues of healthy people were taken as controls.
[0048] 2. Method
[0049] 2.1 Extraction of total RNA in oral cancer group, paracancerous group and normal group
[0050] Extract the RNA of the oral cancer group and the normal group according to the instructions of Kangwei Century Ultrapure RNA Extraction Kit (UltrapureRNAKit (DNaseI), Cat. No. CW0597), prove the integrity of the RNA by gel electrophoresis, and measure the concentration and purity of the RNA with a nucleic acid protein analyzer .
[0051] Use the Ultrapure RNA Extraction Kit (Cat. No. CW0597) to extract, and the main steps are as follows:
[0052] 1. Sample processing, 30-50mg tissue is fully gro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com