H7N9 subtype avian influenza virus detection kit, and application method thereof
A technology for detecting kits and influenza viruses, applied in the field of RT-PCR-ELISA kits, can solve the problems of time-consuming and laborious, expensive test instruments and reagents, and high technical requirements, and achieve easy operation, low cost, and high throughput. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Embodiment 1: Target gene plasmid construction
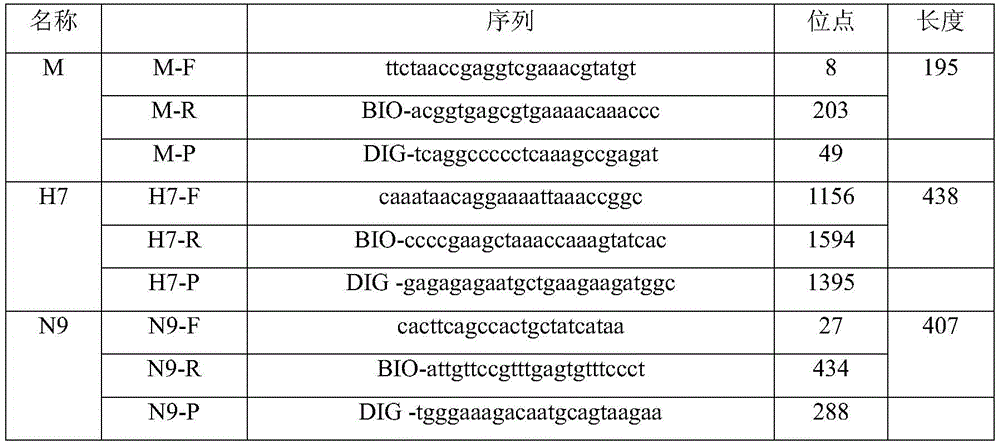
[0017] Design specific primers for the M gene of the influenza A virus, the HA gene of the H7 subtype virus and the NA gene of the N9 subtype virus, and obtain 3 target gene fragments by RT-PCR method amplification, and recover and purify the respective T vectors Connect to obtain target gene positive plasmids, named pMD-M, Pmd-H7 and Pmd-N9 respectively.
Embodiment 2
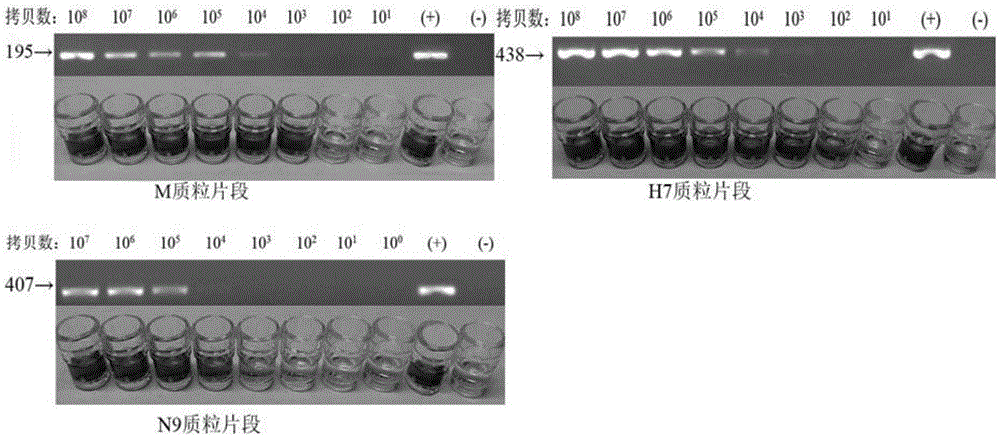
[0018] Embodiment 2: PCR-ELISA method sensitivity
[0019] (1) PCR reaction: Dilute the target gene cloning plasmids pMD-M, Pmd-H7 and Pmd-N9 by 100 times respectively, and then dilute them into 10 times by 10 times. -2 -10 -9 A total of eight concentrations were used as templates, and the above biotin-labeled primers were used to perform PCR amplification reactions using the one-step kit from TaKaRa Company. The reaction conditions are: pre-denaturation at 94°C for 5 minutes, thermal cycle parameters are denaturation at 94°C for 30s, annealing at 56°C for 30s, extension at 72°C for 90s, and after 35 cycles, extension at 72°C for 7 minutes to obtain a digoxin-labeled PCR product. Blank control group. The obtained digoxin-labeled PCR products were determined by ELISA and 1.5% agarose gel electrophoresis to determine their respective sensitivities.
[0020] (2) ELISA detection: Take 5 μL of the amplified biotin-labeled PCR product, add it to 10 μL of 0.1mol / L NaOH solution, d...
Embodiment 3
[0024] Embodiment 3: PCR-ELISA method specificity
[0025] Using enterovirus, respiratory coronavirus, rhinovirus, respiratory syncytial virus, influenza A virus H1, H3, H5, H7 and H9 subtypes and influenza B nucleic acid as templates, PCR-ELISA experiments were performed with the above three sets of primers . Results The 4 sets of primers could not amplify the nucleic acid of enterovirus, respiratory coronavirus, rhinovirus, and respiratory syncytial virus, and there was no cross-reaction between different subtypes of influenza viruses, indicating that the established PCR-ELISA method had good specificity. sex. .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com