Method for identifying human HOTAIR (HOX antisense intergenic RNA) gene polymorphism rs920778 by using MspI
A technology for gene polymorphism and human identification, which is applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of difficulty in popularization and use, expensive restriction endonucleases, and high experimental costs, so as to achieve easy identification, Overcoming the effect of small choice and accurate and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
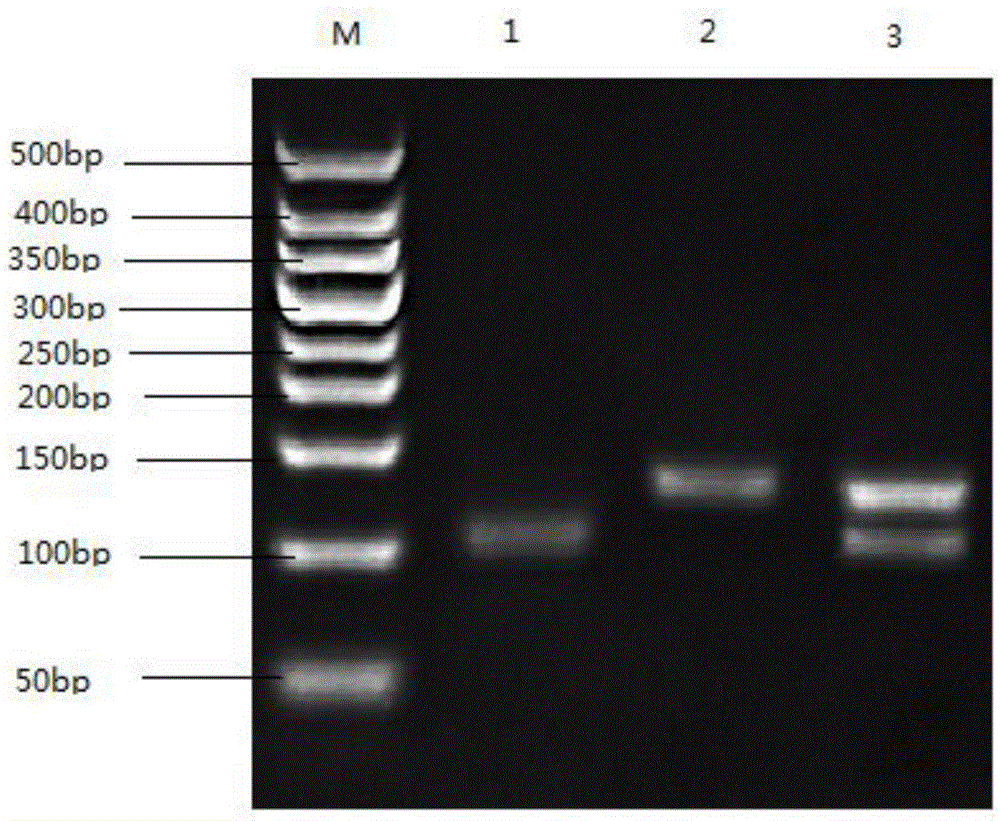
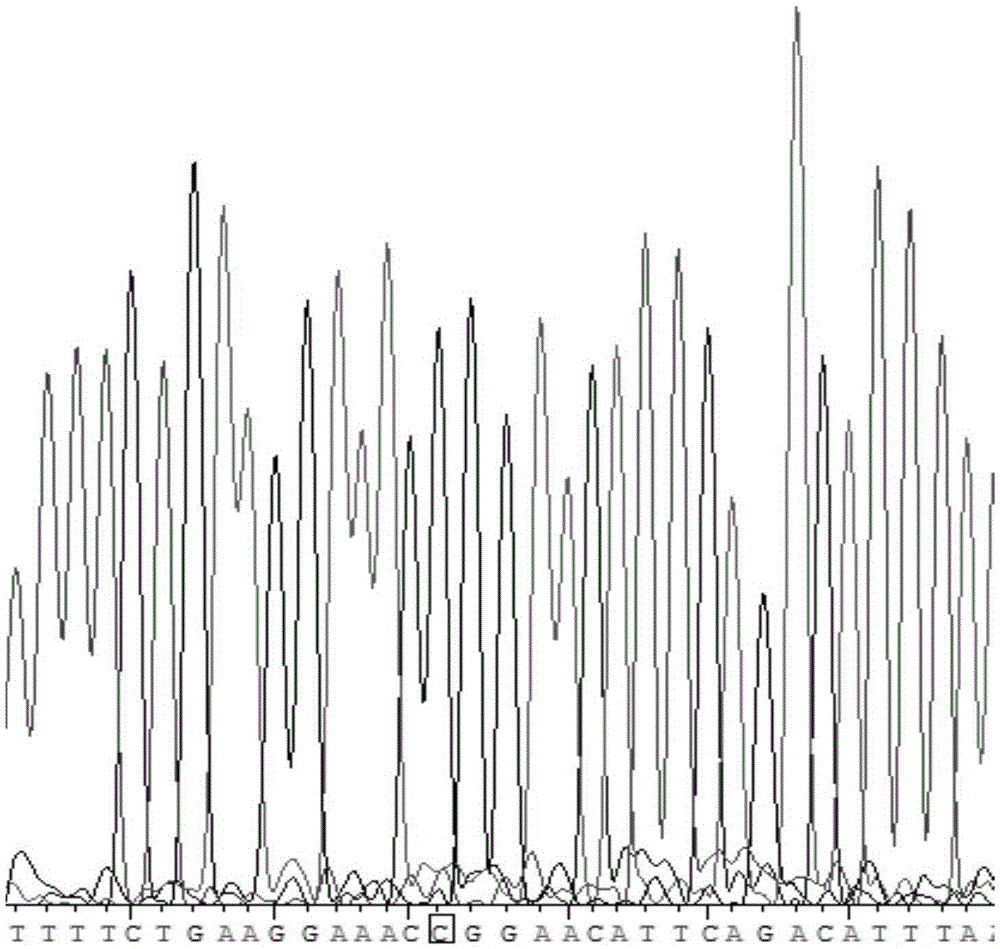
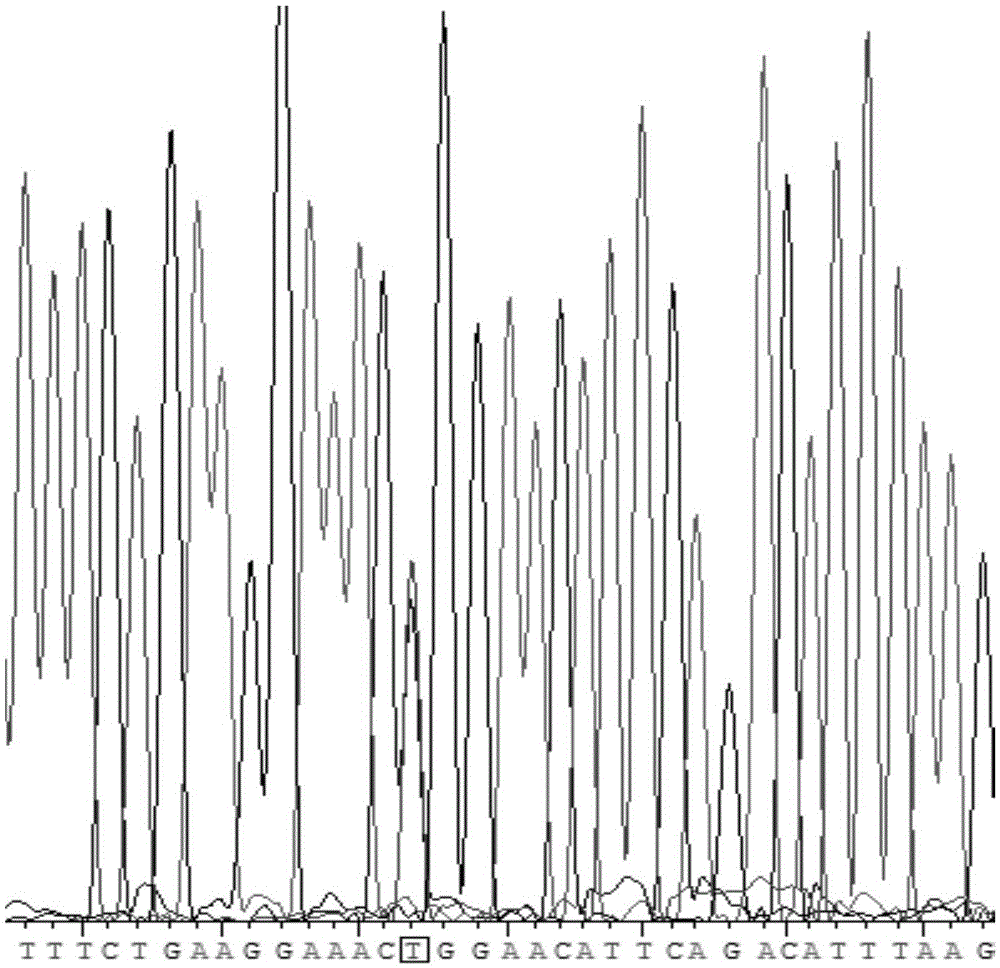
[0055] Example 1. Determination of Human HOTAIRrs920778 Polymorphism in Whole Blood Specimen of Human Peripheral Blood
[0056] 1 Materials and methods
[0057] 1.1 Main instruments and reagents
[0058] Instruments: BCD-228CH Refrigerator (Xinfei Electric), HH-2 Digital Display Constant Temperature Water Bath (Huafon Instruments), SmartGe Gel Imager (Beijing Saizhi Venture), GT9612 Gradient PCR Instrument (Biteke Biology), WD900SL23- 2 models of microwave ovens (Galanz Electric Appliances), DG-300C electrophoresis apparatus (Dingguo Changsheng Biology), etc.
[0059] Reagents: NEP004-1 DNA Extraction Kit (Dingguo Changsheng Biology), 50bpDNALadder (Laifeng Biology), 2×TaqPCRMix (Laifeng Biology), MspI (Thermo), Agarose (SIGMA), etc.
[0060] 1.2 Primer design
[0061] In the dbSNP database of NCBI, the nucleotide sequence of HOTAIRrs920778 was searched. In the primer design software PrimerPremier6.0, parameters such as the length of the primer and the length of the amplifi...
Embodiment 2
[0086] Example 2. Determination of Human HOTAIRrs920778 Polymorphism in Human Breast Cancer Tissue Specimens
[0087] The steps are basically the same as in Example 1, except that the following method is used to extract DNA from breast cancer tissue samples as the DNA to be tested.
[0088] The breast cancer tissue was excised with a mobile phone, and the genomic DNA of the breast cancer tissue was extracted by the phenol-chloroform method as the DNA to be tested.
[0089] 1) Thaw the breast cancer tissue block, wash away the blood stains with normal saline, cut about 0.1g of the tissue and grind it, add 1ml of sterilized water, mix by inversion, centrifuge at 10,000 rpm for 10 minutes, and discard the supernatant. There should be sediment at the bottom of the tube. repeat twice
[0090] 2) Add 200ul of DNA lysate, mix well with 5ul of proteinase K, and digest at 55 overnight.
[0091] 3) After the digestion is completed, add an equal volume of phenol-chloroform mixture (1:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com