Kit for detecting carbapenem-resistant Enterobacteriaceae (CRE) and detection method
A technology of enterobacteriaceae and carbapenems, applied in the field of detection of carbapenem-resistant enterobacteriaceae, can solve the problems of high detection cost, low detection throughput, and poor integration level, and achieve simple operation, good effect, and good Effects of Specificity and Efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Embodiment 1 is used for the preparation of carbapenem-resistant Enterobacteriaceae (CRE) detection kit
[0076] The kit for detecting carbapenem-resistant Enterobacteriaceae (CRE) provided by the present invention consists of the following:
[0077] 1. PCR reaction mixture
[0078] The components and concentrations of the PCR reaction mixture are as follows: 8 μl of Buffer10×buffer solution, pair of detection primers, 0.3 μM of detection probe, and 0.8 mM of each component of dNTP;
[0079] The upstream and downstream detection primers in the detection primer pair are 0.3 μM respectively.
[0080] Detection primer pairs include all of the following detection primer pairs:
[0081] Detection primer pair 1:
[0082] KPC upstream primer is: 5'-ATCGCCGTCTAGTTCTGCTG-3', as shown in SEQ ID NO.1,
[0083] KPC downstream primer is: 5'-TATCCATCGCGTACACACCG-3', as shown in SEQ ID NO.2;
[0084] Detection primer pair 2:
[0085] NDM upstream primer is: 5'-GCAAATGGAAACTGGCGA...
Embodiment 2
[0122] Embodiment 2 is used to detect the use method of carbapenem-resistant Enterobacteriaceae (CRE) kit
[0123] 1. Reaction system preparation
[0124] components
volume
PCR reaction mix
34.4μL
Taq enzyme
0.6μL
sample DNA
5μL
total capacity
40μL
[0125] Take 34.4 μL of PCR reaction mixture, 0.6 μL of Taq enzyme and 5 μL of sample DNA and mix to form 40 μL of reaction solution, vortex and shake evenly, and then centrifuge.
[0126] 2. Fluorescence real-time quantitative PCR reaction and detection
[0127]
[0128] The fluorescent quantitative PCR reaction conditions are shown in the above table: the first stage: 94°C for 5 minutes; the second stage: 94°C for 20s, 58°C for 30s, 40 cycles, and the FAM / HEX signal was collected.
[0129] 3. Result judgment
[0130] After the reaction is over, the amplification curve and cycle number Ct of the samples in each reaction chamber are used as the basis for judging...
Embodiment 3
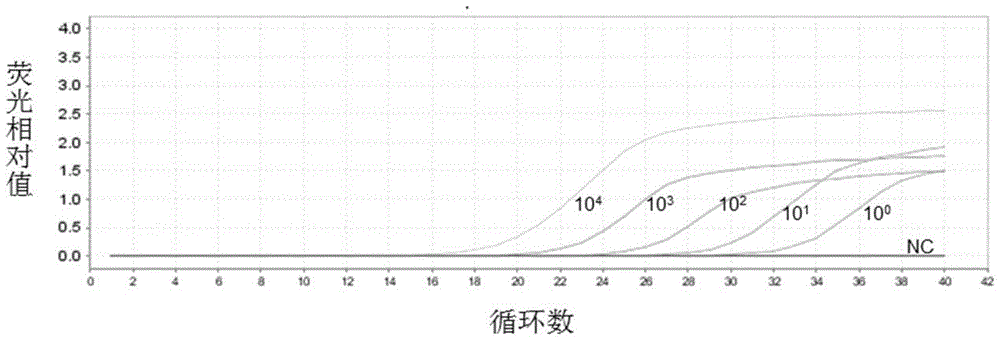
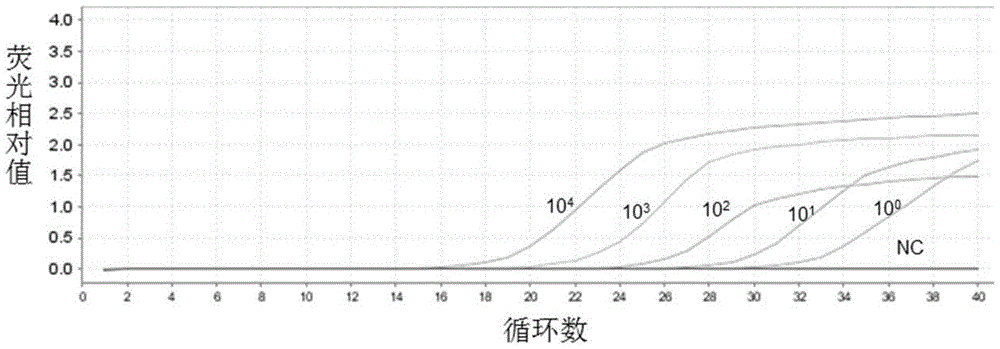
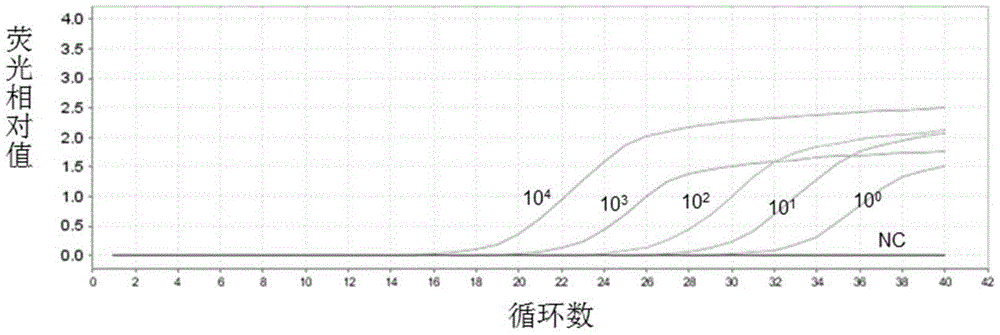
[0132] Example 3. Sensitivity and specificity analysis for detecting carbapenem-resistant Enterobacteriaceae (CRE) kit
[0133] 1. Preparation of reference DNA nucleic acid
[0134] Construct plasmids containing KPC, NDM, IMP, VIM or OXA-48 genes respectively
[0135] Using KPC, NDM, IMP, VIM or OXA-48 to be tested as templates, each standard product (SEQ ID NO.16-SEQ ID NO.20) as described in the summary of the invention was synthesized by Nanjing GenScript Biotechnology Company. Each standard was connected to the pUC57 vector and sequenced for proofreading.
[0136] The recombinant plasmid containing the KPC gene was named pUC57-PKC; the recombinant plasmid containing the NDM gene was named pUC57-NDM; the recombinant plasmid containing the IMP gene was named pUC57-IMP; the recombinant plasmid containing the VIM gene was named pUC57-VIM; The recombinant plasmid of -48 gene was named pUC57-OXA-48.
[0137] 2. Sensitivity and specificity analysis of the kit for detecting car...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com