Sequencing data processing method and processing device
A processing method and processing device technology, applied in the direction of electrical digital data processing, special data processing applications, sequence analysis, etc., can solve the problems of convenience, high throughput, large sample demand, and long time consumption, and achieve the elimination of dependence performance, speed up analysis, and avoid data fluctuations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1 Processing method of sequencing data of samples to be tested
[0062] (1) High-throughput sequencing of cell-free DNA fragments in the peripheral blood of pregnant women to be tested
[0063] (1) Collect the whole blood of pregnant women, and obtain plasma after pretreatment;
[0064] After obtaining the consent notice, 5-10ml of blood was collected from a woman at 22 weeks of pregnancy (i.e. sample S001 in the follow-up table 2) by venipuncture, and added to an ethylenediaminetetraacetic acid (EDTA) tube, and the blood sample was centrifuged at a high speed Afterwards, plasma samples were obtained from which blood cells were removed, and the plasma volume of each sample was about 700ul.
[0065] (2) extract plasma DNA;
[0066] The DNA in the plasma was extracted using the DNA extraction kit HiPure Circulating DNA Kits produced by Magen (Product No. D3180-02).
[0067] (3) Prepare the DNA extracted from plasma into a library that can be sequenced by a n...
Embodiment 2
[0095] Embodiment 2 effectiveness evaluation
[0096] (1) Evaluation using online data samples
[0097] The steps in the processing method shown in Embodiment 1 can be implemented by a computing device in the form of modules or units. In order to evaluate the effectiveness of the method in Example 1, a test is carried out below using a processing device formed of a module or unit capable of performing the above steps. The processing unit includes:
[0098] The sequencing module is used to obtain the nucleotide sequence information of all chromosomes derived from maternal peripheral blood samples through high-throughput sequencing;
[0099] Optionally, the above modules include Illumina's cBot instrument, Illumina's Genome Analyzer, HiSeq2000 / 2500, Hiseq3000 / 4000, NextseqCN500 and other supporting model sequencers, or LifeTechnologies' SOLiD and other matching sequencer modules that perform sequencing functions.
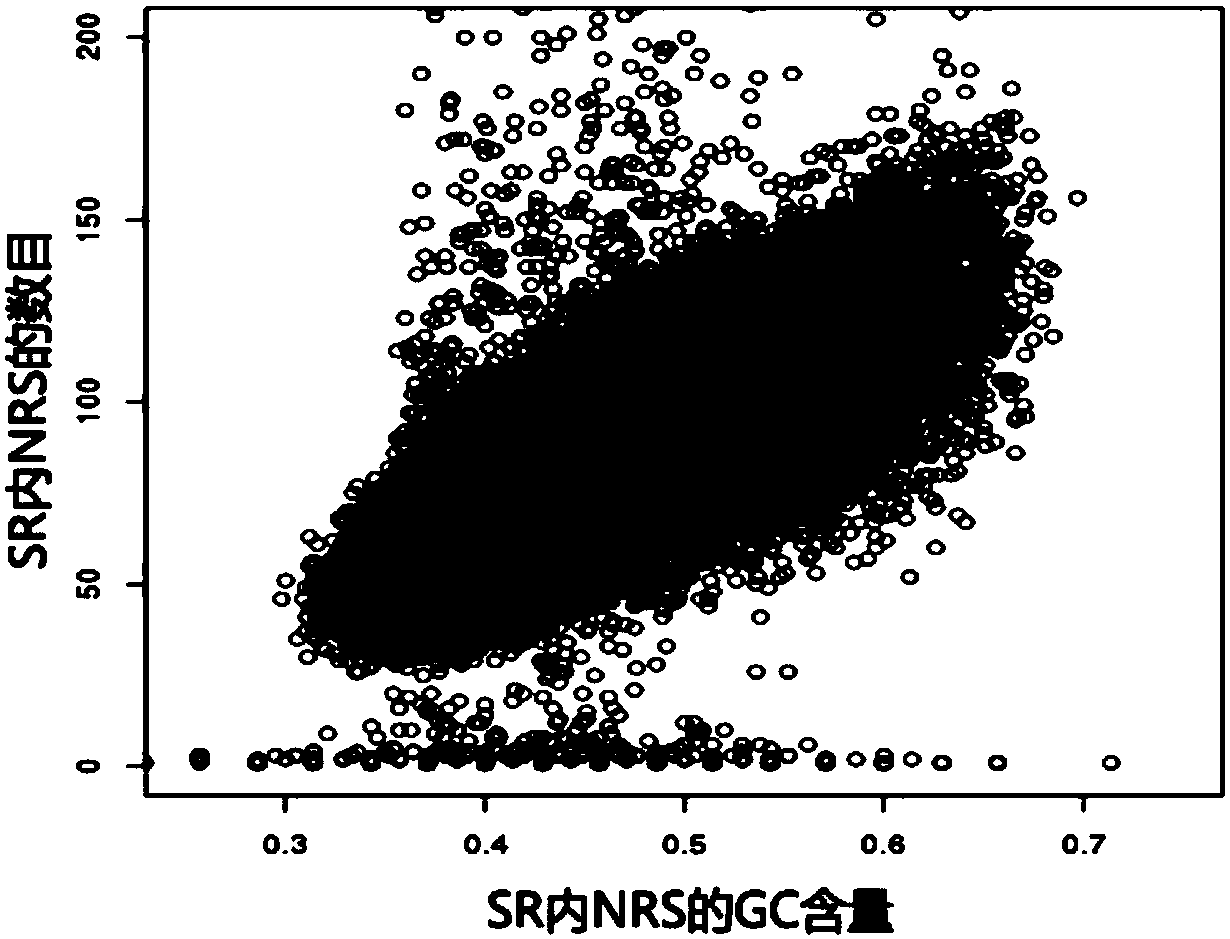
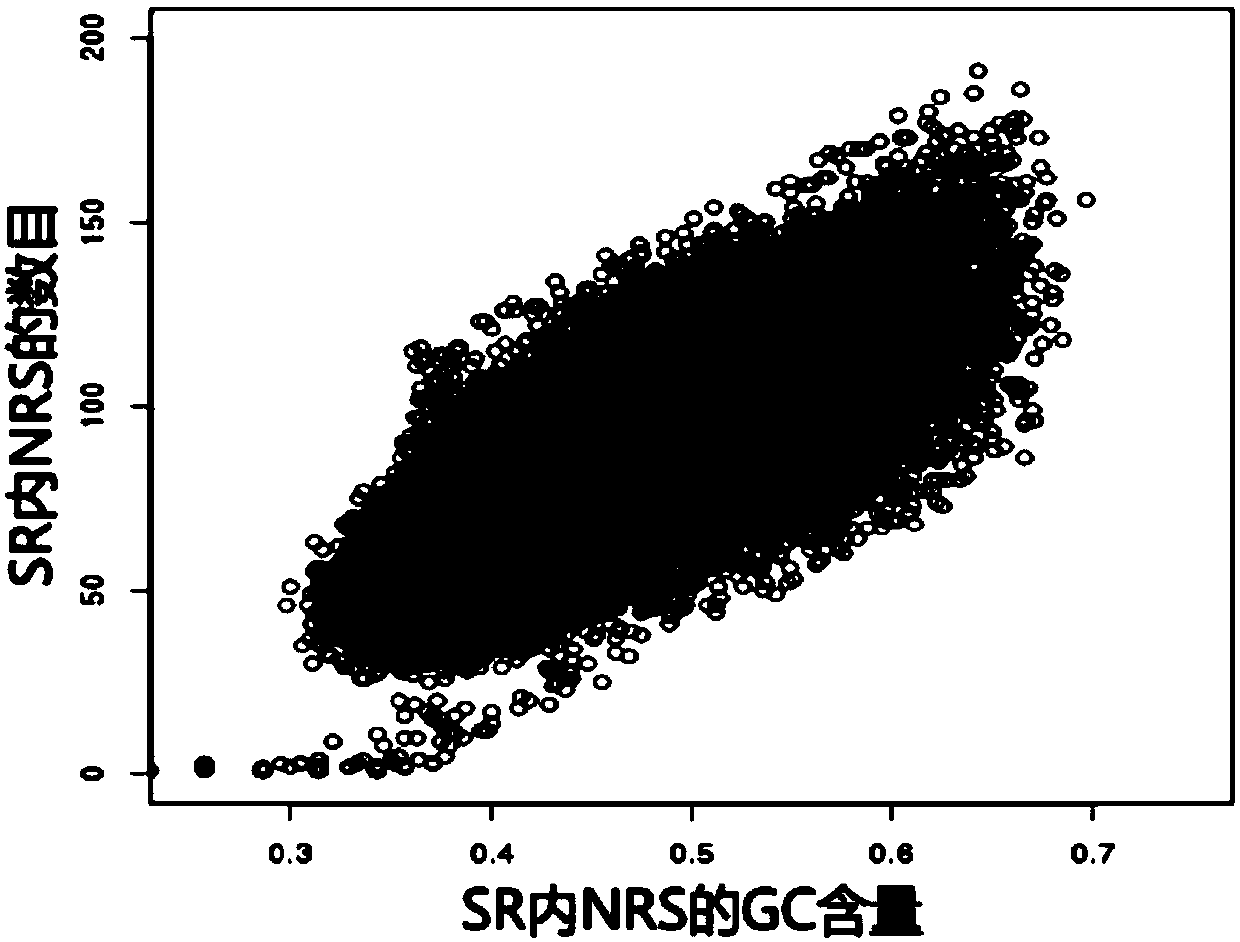
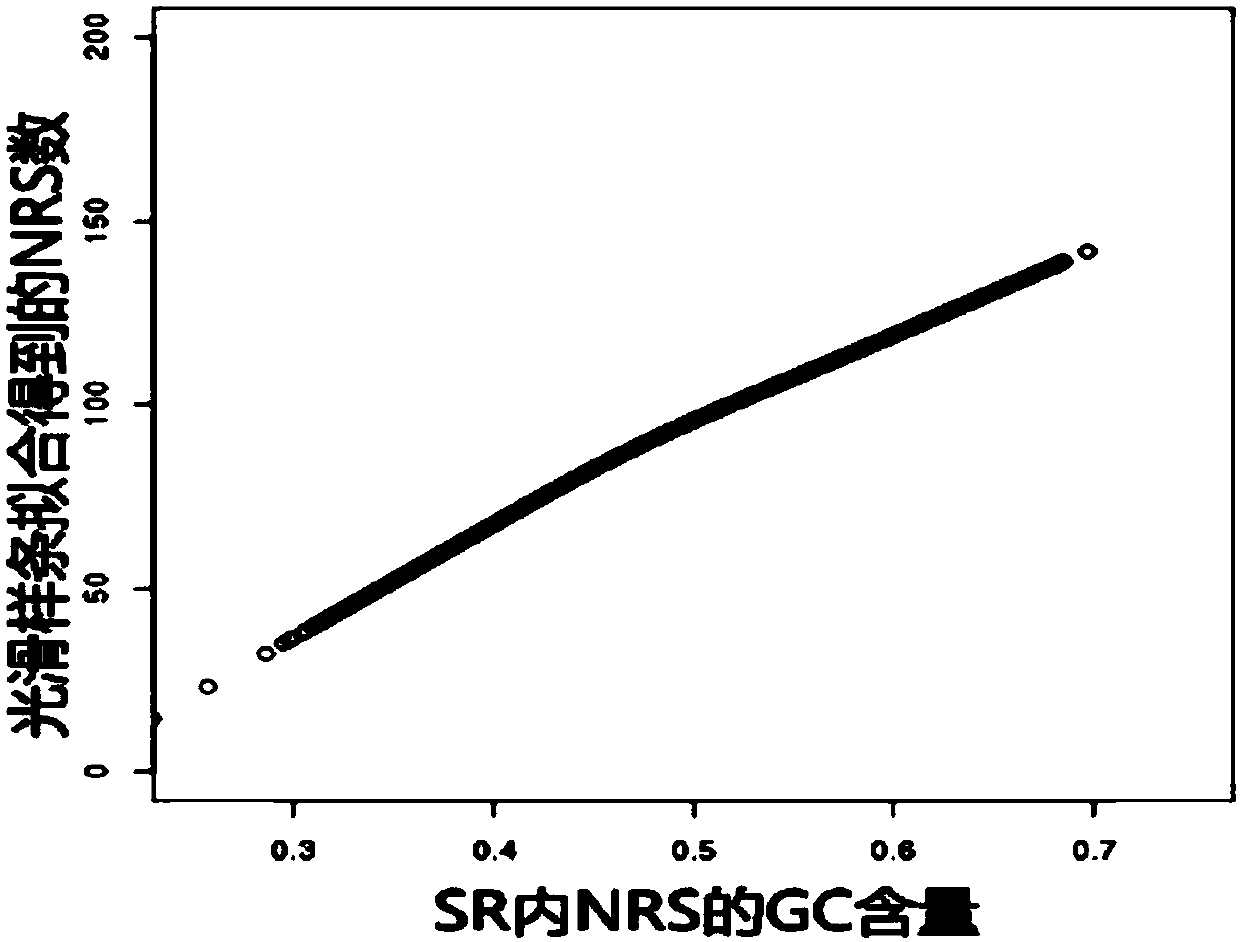
[0100] The specific region division module calls the specific...
Embodiment 3
[0122] Embodiment 3 Stability and Data Volume Research
[0123] (1) Sample stability
[0124] Using the above method, the four samples of s002, s006, s007, and s008 (the corresponding karyotype results are respectively T13 positive, T21 positive, T18 positive and normal) were repeatedly measured 8 times, and the relative chromosome expression was counted (denoted as CR ) and Z test value (denoted as ZV) to study data fluctuations to evaluate the stability of the detection method, and the evaluation results are shown in Table 3.
[0125]
[0126] Table 3. s002, s006, s007, s008 repeatability test data summary table
[0127]
[0128]
[0129] In Table 3 above, Mean represents the mean value, SD represents the standard deviation, and CV represents the dispersion coefficient. It can be seen from Table 3 that the CV (discrete value) of the CR value corresponding to the 4 samples repeated detection 8 times is less than 0.01, and the fluctuation (SD value) of ZV is also wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com