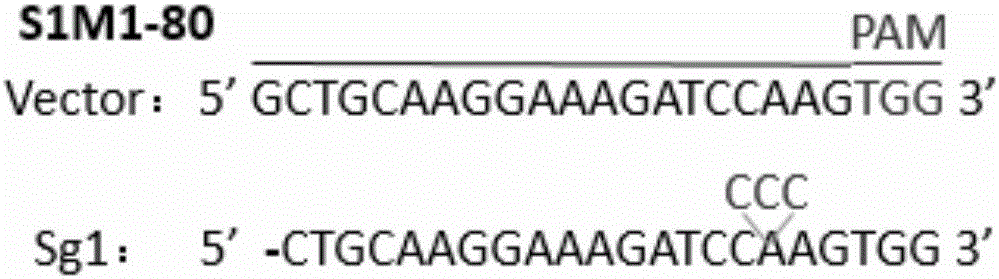SgRNA directing sequence for specific targeting of human ABCG2 gene and application thereof
A kind of specific and genetic technology, applied in the field of sgRNA guide sequence specifically targeting human ABCG2 gene
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
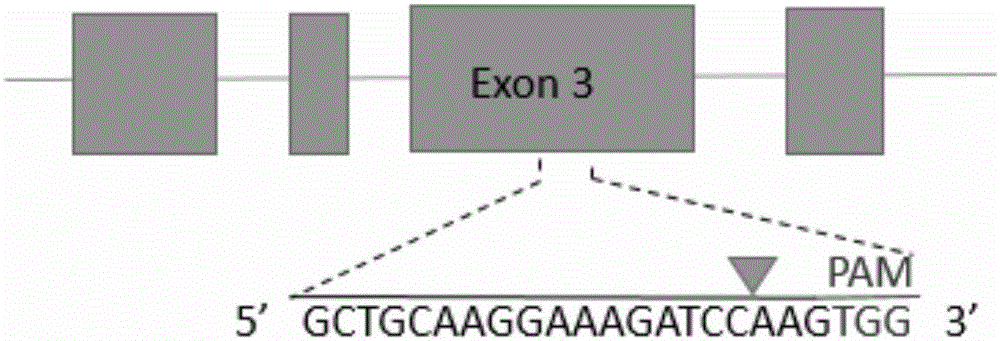
[0037] (1) sgRNA design
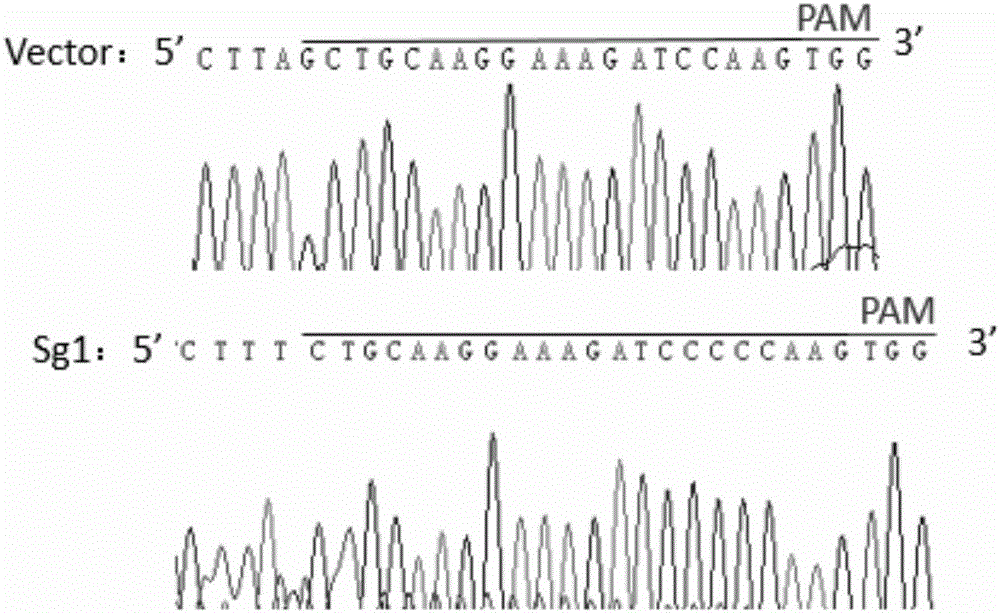
[0038] According to the genome sequence of human ABCG2 gene (gene ID: 9429), a sgRNA targeting human ABCG2 gene was designed. The 20nt oligonucleotide sgRNA guide sequence is: Sg1: 5'-GCTGCAAGGAAAGATCCAAG-3' (located in the third exon of gene ABCG2) ( figure 1 ); add CACCG to its 5' end to obtain a forward oligonucleotide (Forward oligo) (see Sg1-F in Table 1); obtain its corresponding DNA complementary strand according to the guide sequence, and add Add C to AAAC and its 3' end to obtain a reverse oligonucleotide (Reverse oligo) (see Sg1-R in Table 1). Synthesize the above-mentioned forward oligonucleotides and reverse oligonucleotides respectively, denature the Forward oligo and Reverse oligo of the synthesized sgRNA oligonucleotides in pairs, and anneal; At the same time, the designed gRNA target sequence was blasted to exclude non-specific target cutting sites. The specific oligonucleotide sequence is shown in Table 1.
[0039] Table 1 Oligonuc...
Embodiment 2
[0046] (1) Co-transfection of core plasmid and packaging plasmid pMD2.G and psPAX2 into 293T cells
[0047] Cultivate 293T cells until the confluence rate of 293T cells reaches 50% to 60%, 12 to 18 hours after seeding is the best time for transfection; replace fresh culture medium before transfection, add 3mL medium to a 60mm dish; The amount used is 4 μg of core plasmid (lentiCRISPRv2-hABCG2-Sg1 prepared in Example 1, and lentiCRISPRv2 empty vector as a control), 3 μg of psPAX2, 1 μg of pMD2.G, 24 μL of PEI, supplemented with DMEM to a total volume of 200 μL, and the order of addition DMEM, PEI, and plasmid DNA (core plasmid, pMD2.G, and psPAX2) respectively; then stand at room temperature for 30 minutes to fully polymerize PEI and plasmid DNA, and add the transfection system dropwise to the above-mentioned small dish with 293T cells after polymerization , shake gently and place in 37°C, 5% CO 2 Continue culturing in the incubator;
[0048] (2) Virus harvest and concentrati...
Embodiment 3
[0061] Example 3 In vitro cell experiments to identify the knockout effect of the human gene ABCG2 on the cells after screening
[0062] (1) western blot
[0063] The ABCG2 protein expression level of the cell line S1M1-80sg1, which successfully knocked out the ABCG2 gene in Example 2, was identified by western blot experiments. The cell line S1M1-80 successfully transfected with the empty vector lentiCRISPRv2 was used as a blank control, S1 was used as a positive control, and S1M1-80 was used as a positive control. 80 as a negative control; among them, the primary antibody is Anti-ABCG2 (MM SC-58222, purchased from santacruze), the secondary antibody is Anti-mouse IgG, HRP-linked Antibody (product number 7076, cell signaling), and the specific method is conventional western blot operation process.
[0064] The result is as Figure 4 As shown, in S1M1-80 cells, there is a significant difference in the expression of ABCG2 between the cells transformed into sg1 and the cells o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com