Rapid identification method for present generation genotype of gramineous seed
A kind of undergraduate and seed technology, applied in the fields of molecular biology and genetic breeding, can solve the problems of increasing screening costs, complicated operation procedures, and long time consumption, and achieve the effects of saving manpower, safe operation, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1 seed treatment
[0039] The specific method is as follows:
[0040] 1. Hull removal: remove the glume of a single seed;
[0041]2. Cut the endosperm: After shelling the single seed, cut the endosperm horizontally at 1 / 3-1 / 2 of the seed with a knife, put the endosperm in a 2mL centrifuge tube, and put the part containing the complete embryo in another centrifuge tube, corresponding to serial number, store dry at room temperature;
[0042] 3. Soaking: add 0.5-1mL distilled water to the endosperm in step 2), soak at room temperature for 3 days.
[0043] Comparative example 1 seed treatment
[0044] The specific method is as follows:
[0045] 1. Hull removal: remove the glume of a single seed;
[0046] 2. Cut the endosperm: After shelling the single seed, cut the endosperm horizontally at 1 / 3-1 / 2 of the seed with a knife, put the endosperm in a 2mL centrifuge tube, and put the part containing the complete embryo in another centrifuge tube, corresponding to...
Embodiment 2
[0058] Example 2 Seed Endosperm DNA Extraction
[0059] The specific method is as follows:
[0060] 1. Crushing: Drain the differently treated endosperms with distilled water, add 1 mL of 2×CTAB solution, add a small steel ball with a diameter of 5-6 mm, and then crush them on a fast sample preparation machine (FastPrep-24 from MP Company). Start 30sec each time, break 2-4 times in total;
[0061] 2. Water bath: put the crushed endosperm in a water bath for water bath, 65°C, 30-60min, and mix several times during the period;
[0062] 3. Centrifugation: Centrifuge the extract after the water bath, 12000rpm, 15min, take 0.8mL supernatant into a new centrifuge tube;
[0063] 4. Precipitation: Add 0.8 times the volume of isopropanol to the supernatant, mix it upside down, store at low temperature (-20°C) for 10 minutes; centrifuge at 12,000 rpm for 10 minutes at 4°C, discard the supernatant, and keep the precipitate;
[0064] 5. Washing: add 0.5mL of 75% ethanol to the precipit...
Embodiment 3
[0067] Example 3 Genomic DNA Quality Identification
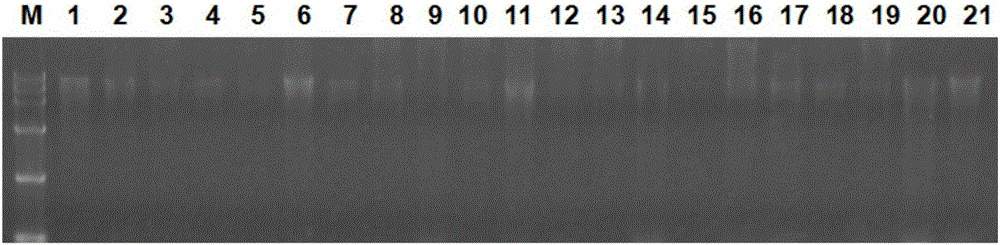
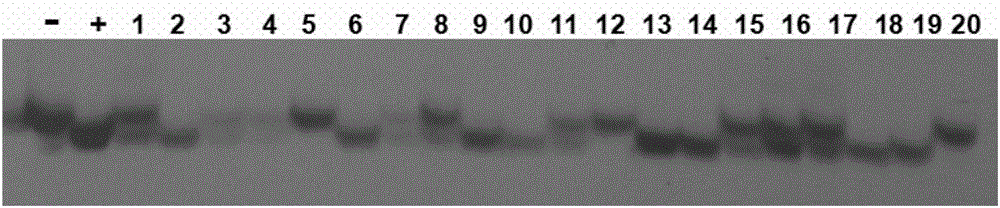
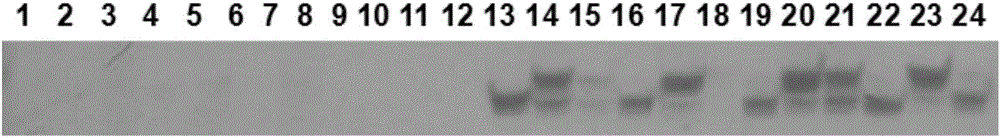
[0068] Pipette 5 μL of the extracted single-grain endosperm DNA solution and 1 μL of loading buffer (China Keruitai) to mix, and then pipette 5 μL of the mixed sample and Maker (China Keruitai) to 1% agarose gel to make Electrophoresis at 350 mA for 25 min. see results figure 1 , the DNA band after treatment 3d is clearly visible, and there are few impurities, which shows that the single-grain endosperm DNA extracted by the method of the present invention has good quality, high concentration and high purity. The DNA of treatment 0-2d had no obvious bands. The DNA concentration of measuring treatment 3d is 110-130ng / μL, and its OD260 / OD280 value is between 1.9-2.0, shows to contain a little RNA sample in the DNA sample extracted by the present invention, and this is relevant with undigested RNA, but does not affect PCR amplification effect.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com