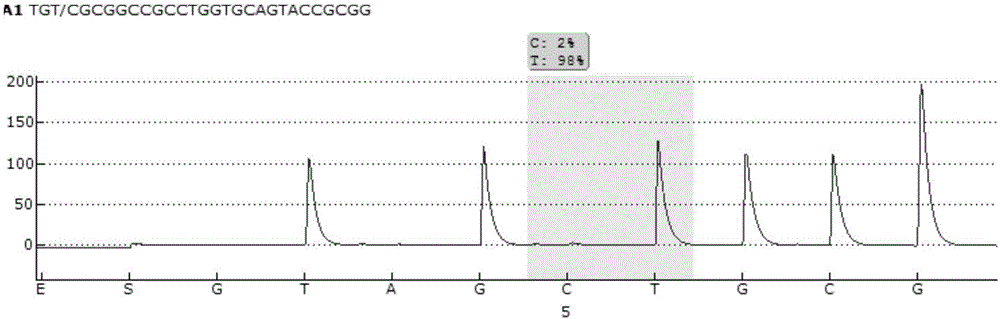
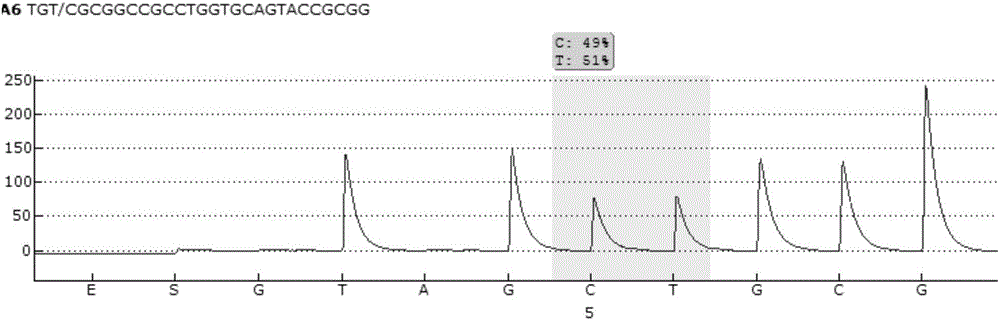
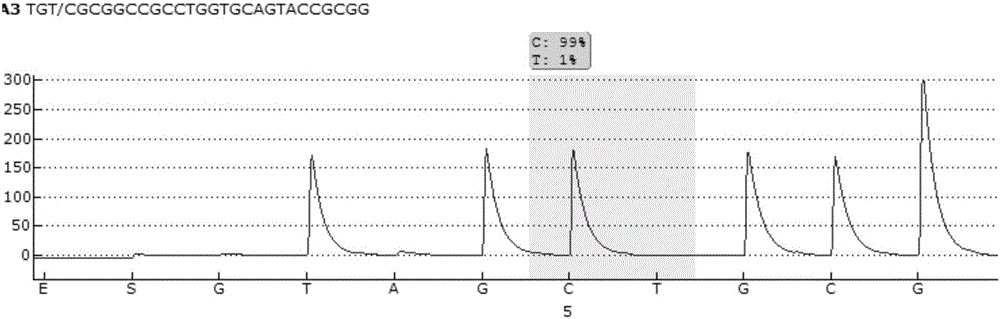
Pyrosequencing primer pair for qualitatively detecting ApoE genetic typing and kit
A technology of pyrosequencing and genotyping, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., and can solve the problems of low sensitivity of ApoE genotyping, long detection cycle, cumbersome operation, etc. problems, to achieve the effects of fast sequencing speed, short reaction time, and simple sample processing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Embodiment 1: the preparation of kit
[0073] 1. Design and synthesis of primers and probes
[0074] For the polymorphic sites Cys112Arg and Arg158Cys alleles of the human ApoE gene, select specific mutation sites, and use the PyroMark Assay Design2.0 software to design primers; the amplification primers and sequencing primers are first purified by PAGE, and then HPLC Purified, wherein the 5' ends of SEQ ID NO.2 and SEQ ID NO.5 were biotin-labeled.
[0075] Table 1. Mutation site and type:
[0076] Mutation
[0077] The amplified sequence is shown in Table 2:
[0078] Table 2. Specific amplification primers and primer sequences
[0079]
[0080]
[0081] 2. Selection of reference substance
[0082] A synthetic oligonucleotide chain TAYGGTTTGCA control oligo was used as the quality control product; DNase / RNase-Free water was used as the blank control product.
[0083] 3. Composition of PCR reaction solution
[0084] Table 3. Composition of PCR react...
Embodiment 2
[0089] Embodiment 2: the use of kit
[0090] 1. Sample testing
[0091] Dissolve the dry powder of the primers (the validity period of the primers is 1 month after dissolution). Prepare the system according to the number of templates: take the PCR reaction solution, add dissolved primers, uracil DNA glycosylase, and Taq DNA polymerase, aliquot the system, add sample DNA, blank control substance or positive control substance as templates to form PCR reaction system. Perform PCR amplification according to the PCR reaction procedure.
[0092] The main components of the ApoE Cys112Arg and ApoE Arg158Cys systems are as follows:
[0093] Table 5. The main components of the ApoE Cys112Arg system
[0094]
[0095] Table 6. Main components of ApoE Arg158Cys system
[0096]
[0097] The system reaction procedure is as follows:
[0098] Table 7. PCR reaction program
[0099]
[0100]
[0101] After the amplification is completed, check the PCR results on agarose gel to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com