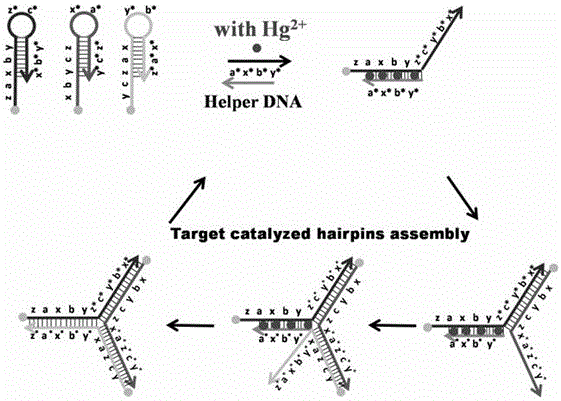
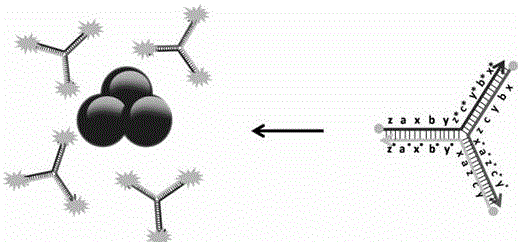
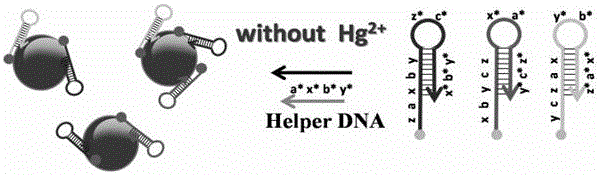Colorimetric and fluorescent double-signal biosensor for detecting Hg<2+>, and detection method of biosensor
A biosensor and detection method technology, applied in the field of biosensing, can solve problems such as low sensitivity and narrow applicability, achieve high amplification efficiency, wide application range, and overcome false positive effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0036] The following examples will further illustrate the present invention, but do not limit the present invention thereby.
[0037] In the following examples:
[0038] Chlorauric acid, sodium citrate, Tris-HCl buffer solution, mercuric nitrate, FAM, and concentrated sulfuric acid are analytically pure, and ultrapure water is self-made in the laboratory. The nucleotide sequences used in Examples 1-4 are shown in Table 1. The number of T mismatches between the helper DNA and H1 in Examples 1-4 is 6.
[0039] Auxiliary DNA and nucleotide sequences used in table 1 embodiment 1-4
[0040]
Embodiment 5
[0041] The nucleotide sequences used in Example 5 are shown in Table 2. The number of T mismatches between the helper DNA and H1 in Example 5 is 8.
[0042] Auxiliary DNA and nucleotide sequence used in table 2 embodiment 5
[0043]
[0044] The sequence listings of HI, H2, and H3 in Tables 1 and 2 above are shown in SEQUENCE LISTING.
[0045] instrument
[0046] Fluorescence photometer, the detection conditions are: the excitation wavelength and emission wavelength of FAM are set to 492nm and 517nm respectively, the slit width is 10nm, the fluorescence emission spectrum of the sample is excited by 492nm light, and the scanning range is: 505-600nm, scanning step The length is 1nm.
[0047] UV-visible spectrophotometer.
[0048] Since the sensitivity of fluorescence detection is higher than that of colorimetric detection, the following examples are all detected with a fluorescence photometer, and only a brief description is given for colorimetric detection.
Embodiment 1
[0050] Preparation of AuNPs
[0051] Weigh 0.01g of chloroauric acid and dissolve it in 100mL of ultrapure water, heat and boil under reflux; add 2.5mL of 1wt% sodium citrate solution under rapid stirring, heat and react for 30min and then cool to room temperature naturally; the prepared AuNPs are kept at 4°C±2°C save in .
[0052] Dual signal Hg with colorimetric and fluorescent signals 2+ Preparation and detection process of biosensor
[0053] Mix the Tris-HCl buffer solution of Hg(NO3)2, molecular hairpin probes H1, H2 and H3 and the Tris-HCl buffer solution of the auxiliary DNA, wherein the concentration of the Tris-HCl buffer solution is 50mmol / L, containing 50mmol / L MgCl2 and 0.5mol / L NaCl, pH 8.0, the volume after mixing is about 100μL, the concentration of molecular hairpin probes H1, H2 and H3 is 30-300nmol / L, the concentration of auxiliary DNA is 20-500nmol / L, Hg2+ The concentration of AuNPs is 0.2-100nmol / L, react at room temperature for 1-5h, then add 900μL AuNP...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Slit width | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com