Method for analyzing otis tarda food source plant through feces DNA (Deoxyribonucleic Acid)
A technology of great bustards and plants, which is applied to the analysis of food source plants of great bustards by fecal DNA and the analysis of food habits of great bustards, can solve the problems of unclear food types and nutritional requirements of great bustards, lack of scientific guidance, and unsuccessful artificial breeding.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0115] Example 1. Analysis of Great Bustard Food Source Plants Based on Chloroplast DNA trnL Barcodes
[0116] (1) Establishment of the trnL complete sequence database of native plant chloroplast DNA
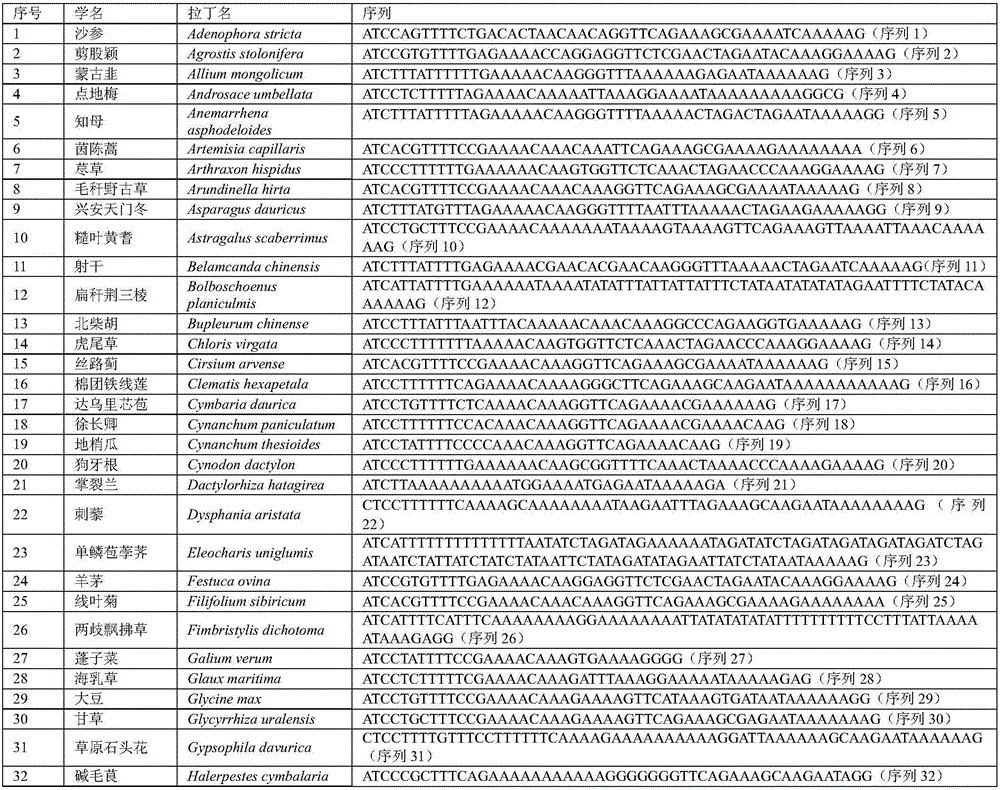
[0117] 1. Collection of potential food plants
[0118] Collect plant leaves at the site of great bustard activity or where there are traces of great bustard activity (feces, feathers, scratches, etc.), put the fresh leaves of the plant into a paper bag, add silica gel, and store in a dry place. Record the family, genus, and species information of the collected plants.
[0119] 2. Plant DNA extraction
[0120] The leaves were put into a mortar, ground into powder with liquid nitrogen, and the plant genomic DNA was extracted using a broad-spectrum plant genomic DNA rapid extraction kit (article number: DL116-01, Beijing Biomed Gene Technology Co., Ltd.).
[0121] Perform agarose electrophoresis on the extracted DNA, and use a UV spectrophotometer to measure the concentration an...
Embodiment 2
[0174] Example 2, method verification based on feces DNA analysis of bustard sex
[0175] (1) Collect known plants to feed the Great Bustard in cages and collect feces
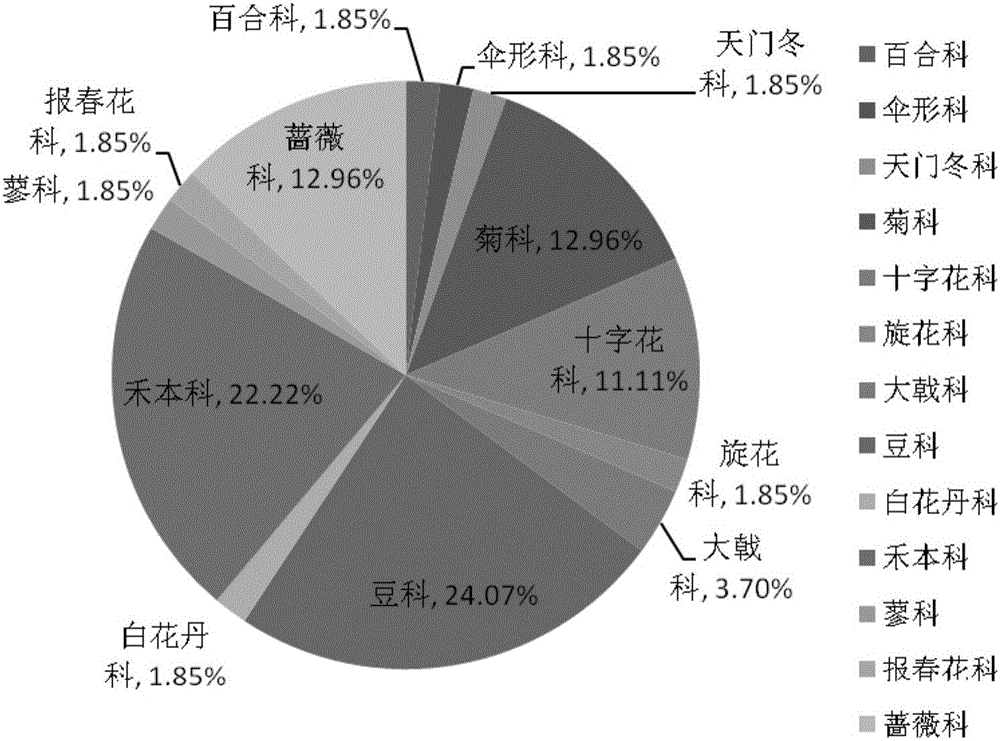
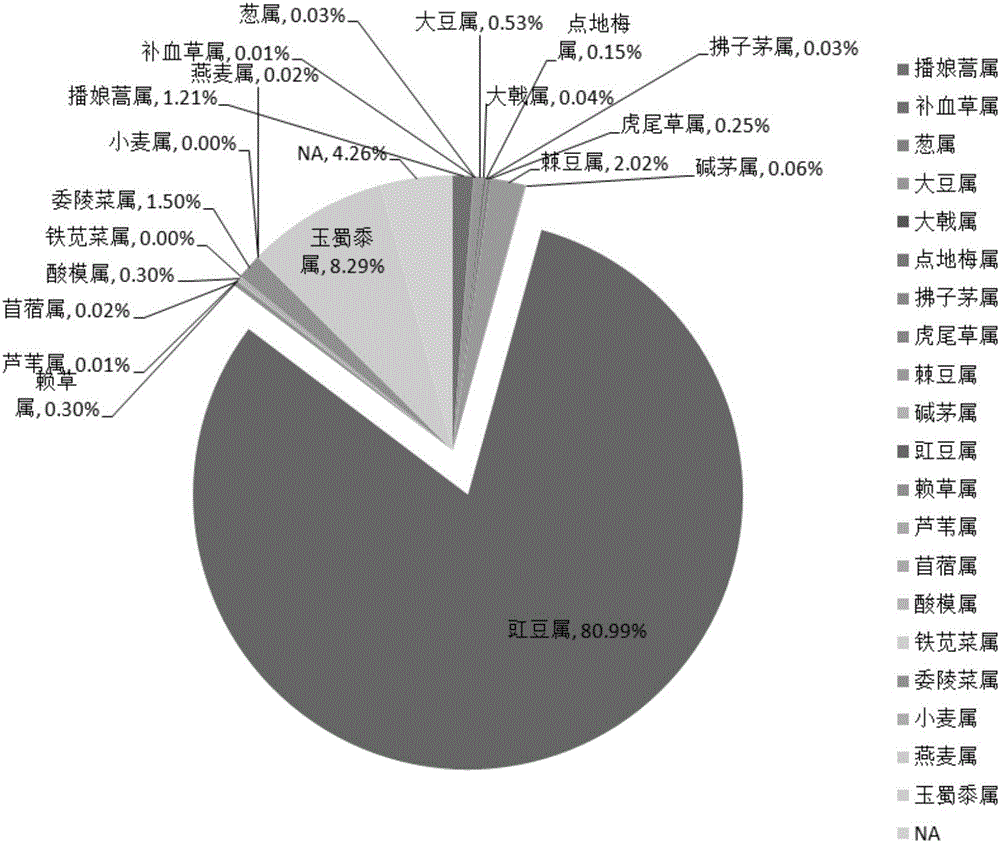
[0176]In places where there are bustard activities or traces of bustard activities in the wild (feces, feathers, scratches, etc.), 10 whole plants of 8 kinds of plants were collected, and the plant species were Mongolian allium (Androsace umbellata) and Diandimei (Anemarrhenaasphodeloides), Bupleurum chinense, Leymus chinensis, Chloris virgata, Rhaponticum uniflorum, Scabiosa comosa, Potentilla discolor. In addition, 2 plant staples, mung beans (Vigna radiata) and maize (Zea mays), were fed.
[0177] Before feeding the caged bustard, remove old feces, dropped food and weeds on the ground in advance to avoid interfering with subsequent analysis. Select 6 adult great bustards with good daily eating conditions as the experimental subjects, and give them enough and clean water during the feeding period. No food...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com