Novel xylanase produced from streptomyces sp. strain hy-14
A xylanase, a typical technology, applied in the direction of enzymes, hydrolases, biochemical equipment and methods, etc., can solve problems such as complex procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1: Isolation and Identification of Streptomyces Strains
[0078] In order to isolate xylanase-producing microorganisms, the present inventors conducted the following experiments to isolate microorganisms having enzymatic activity to hydrolyze xylan from symbiotic intestinal microorganisms of the oriental mole cricket.
[0079] Specifically, the collected oriental mole crickets were soaked in 70% (w / w) ethanol for 1-2 minutes to remove any contaminants on the surface of the insects. Wash the insect carcasses twice with sterile distilled water to remove ethanol. The insects were dissected and the intestines were removed with forceps. Place the intestines in phosphate-buffered saline (0.8% NaCl, 0.02% KCl, 0.144% Na 2 HPO 4 , 0.024% KH 2 PO 4 ) and crushed. To isolate microorganisms capable of hydrolyzing xylan, Difco TM R2A agar (BD) was used as basal medium, and medium supplemented with 2 g / L azoxylan (Megazyme) was used to prepare selective medium. By u...
Embodiment 2
[0082] Embodiment 2: Determination of xylanase gene
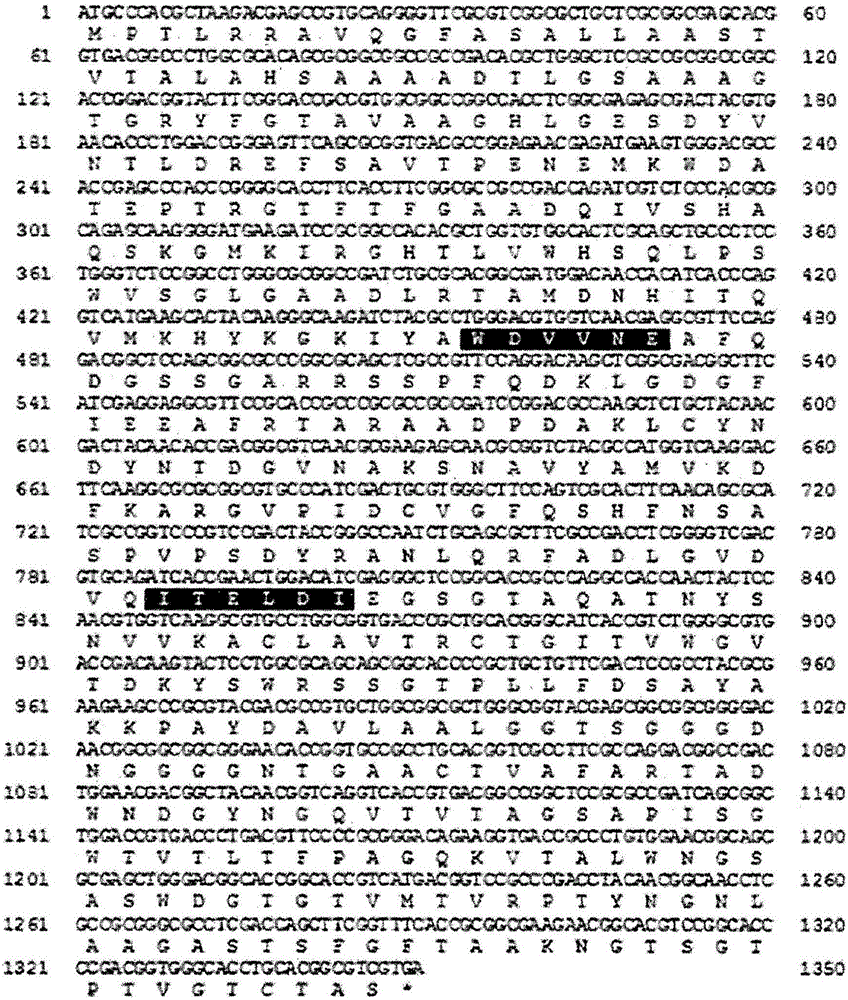
[0083] In order to find the gene capable of degrading xylan of the Streptomyces strain HY-14 (preservation number: KCTC 12531BP) isolated in Example 1, such as figure 1 As shown, by using the primers constructed based on the GH10 (glycoside hydrolase family 10) xylanase gene conservative sequence (WDVVNE, ITELDI) of the genomic DNA of the above-mentioned bacterial strain, the protein (SEQ.ID .NO:1) polynucleotide sequence (SEQ.ID.NO:12) for amplification.
[0084] In particular, genomic DNA was isolated from the above-mentioned strains. PCR was performed by using genomic DNA as a template with: 10× buffer (MgCl 2 ), 2.5mM dNTPs, 5× buffer, FastStart Taq DNA polymerase (Roche) and primers [GF-10 primer: 5'-TGGGACGTCSTCAACGAG-3' (SEQ.ID.NO:6); GR-10 primer: 5 '-GATGTCGAGCTCSGTGAT-3' (SEQ. ID. NO: 7)]. At this time, the PCR conditions were as follows: pre-denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seco...
Embodiment 3
[0086] Embodiment 3: Construction of xylanase recombinant vector
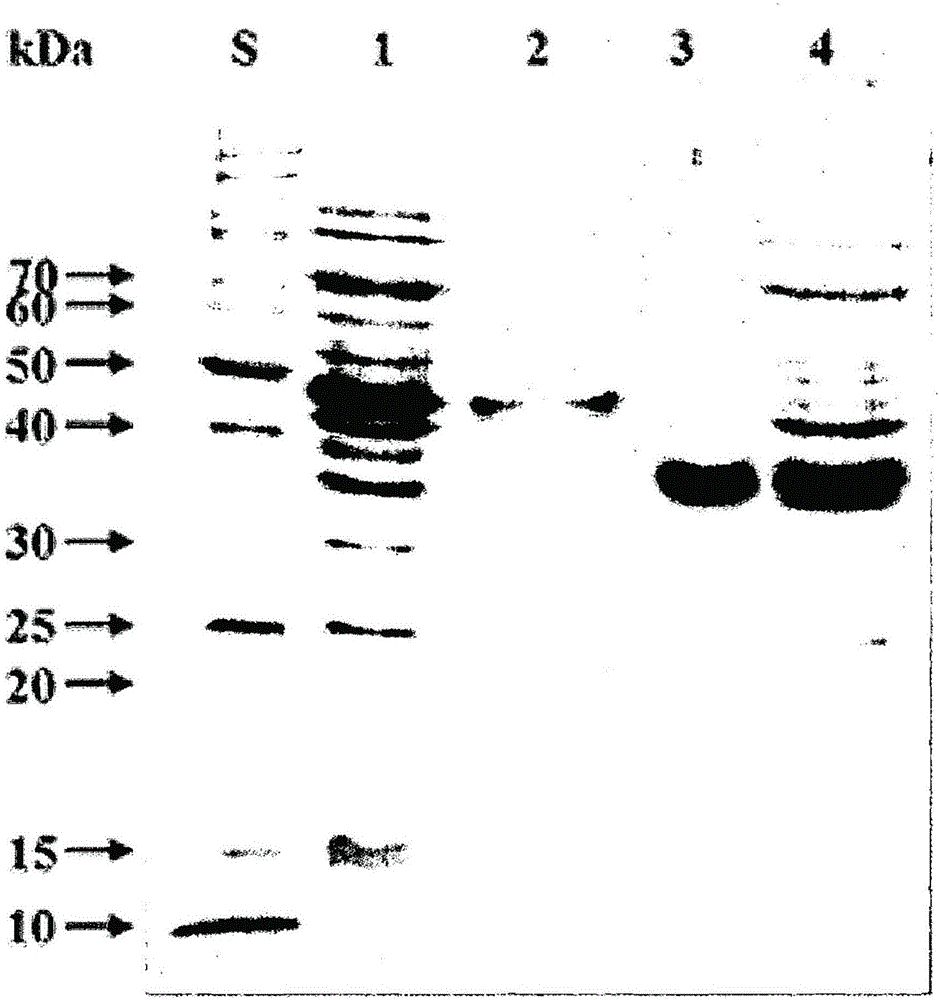
[0087] The following experiments were performed to clone the total XylU and XylUΔCBM2 obtained in Example 2.
[0088] Specifically, the PCR product obtained above was purified by using NucleoSpinGel and PCR Clean-up Kit (Machery-Nagel), followed by ligation in pGEM-T easy vector (Promega, Madison, USA). Escherichia coli DH5α was transfected with the above-mentioned vectors, and then the transformed strains were selected from LB agar plates supplemented with ampicillin. The recombinant pGEM-T vectors were isolated and purified from the selected transformed strains and named "pGEM-T-XylU" and "pGEM-T-XylUΔCBM2". pGEM-T-XylU and pGEM-T-XylUΔCBM2 were digested with NdeI and HindIII, and then XylU and XylUΔCBM2 genes were purified by using NucleoSpinGel and PCR Clean-up Kit (Machery-Nagel). The expression vector pET-28a(+) (Novagen, USA) was additionally digested with NdeI and HindIII, followed by purification. I...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com