Infectious spleen and kidney necrosis virus (ISKNV) gene intron and its use in distinguishing of live and inactivated ISKNVs
An intron, live virus technology, applied in double-stranded DNA viruses, viruses, viruses/phages, etc., to achieve high sensitivity, high sensitivity, and shorten the inactivation test period.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Screening and verification of intron IN-3
[0052] 1. Screening and primer design of intron IN-3
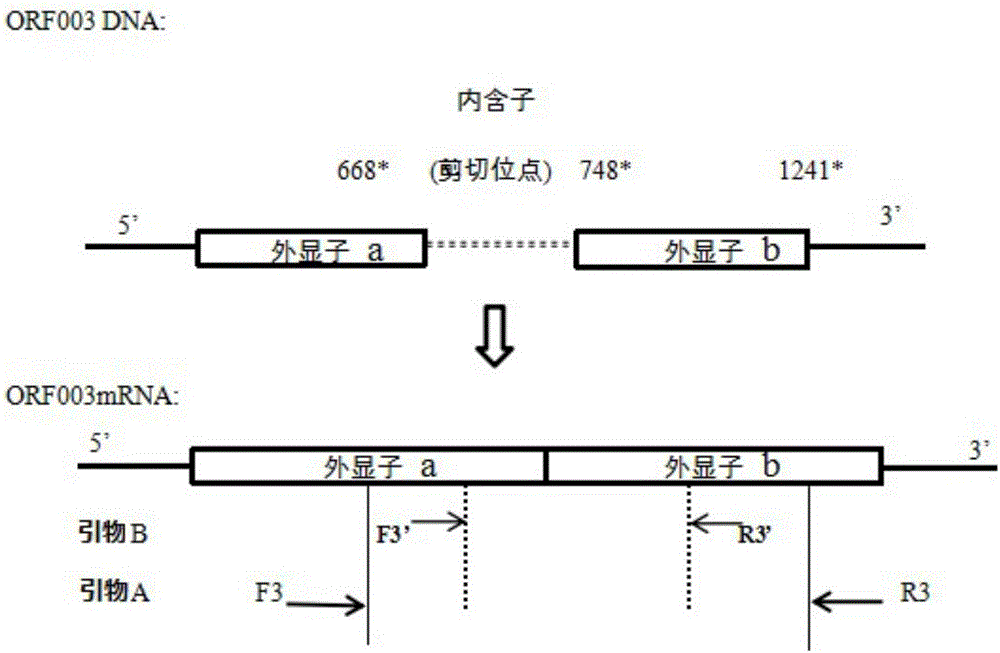
[0053] According to the results of transcriptional profiling of CPB cells infected with ISKNV in our laboratory, compared with the genome sequence of ISKNV (NC_003494.1), we screened out Unigene0020179 (ORF003L) at the position of 668bp~748bp, containing an 80bp inclusion Sub, named intron IN-3, see Table 1.
[0054] Table 1 Genome sequence information where introns are located
[0055]
[0056] Primer 5.0 software was used to design specific primers, and the primer information and expected DNA fragment and cDNA fragment sizes are shown in Table 2.
[0057] Table 2 List of PCR primers designed across introns
[0058]
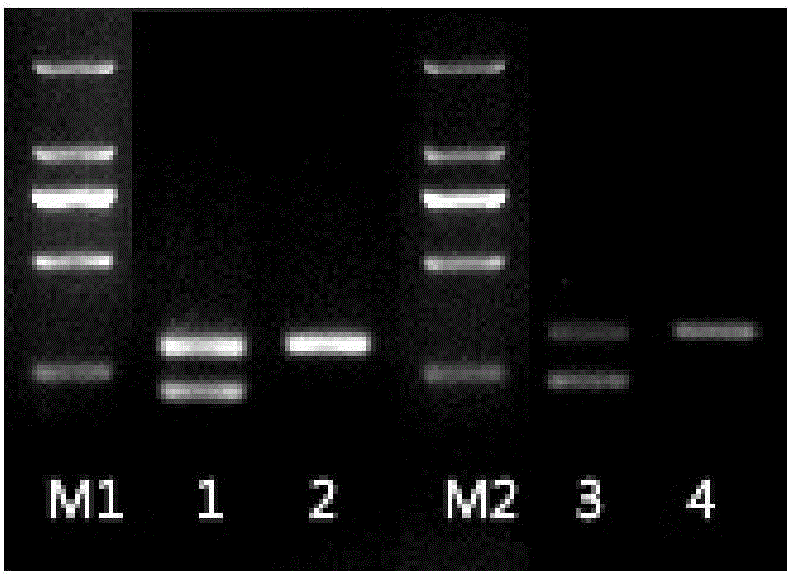
[0059] 2. Intron verification
[0060] 1. RNA extraction and DNA positive template preparation
[0061] Dilute ISKNV virus stock solution 10×, 100×, inoculate CPB cells and incubate for 72 hours, then extract total RNA, each T25cm 2 The...
Embodiment 2
[0070] Embodiment 2 Nested PCR amplification
[0071] According to ISKNV, the ORF003L gene contains an intron IN-3 located at the position of 668bp-748bp and the size is 80bp. like figure 2 As shown, after the DNA is transcribed to generate mature mRNA, the intron will be cleaved, and nested primers (Table 3) are designed on exon a and exon b, respectively, for PCR amplification.
[0072] Table 3 is used for the IN-3 gene primer of nested PCR amplification
[0073]
[0074] The 25 μl reaction system used for the first round of amplification is: 12.5 μL 2×Taq PCR StarMix, 1 μL each of primers F3 and R3 at a concentration of 10 μM, 1 μL DNA or cDNA template, 9.5 μL ddH 2 O; Reaction conditions: 94°C for 5min, 94°C for 45s, 68°C for 45s, 72°C for 90s, 30 cycles, 72°C for 10min, the product was detected by 2% agarose electrophoresis.
[0075] The 25 μl reaction system used for the second round of amplification is: 12.5 μL 2×Taq PCR StarMix, 0.5 μL each of primers F3’ and R3...
Embodiment 3
[0077] Embodiment 3 nested PCR method sensitivity test
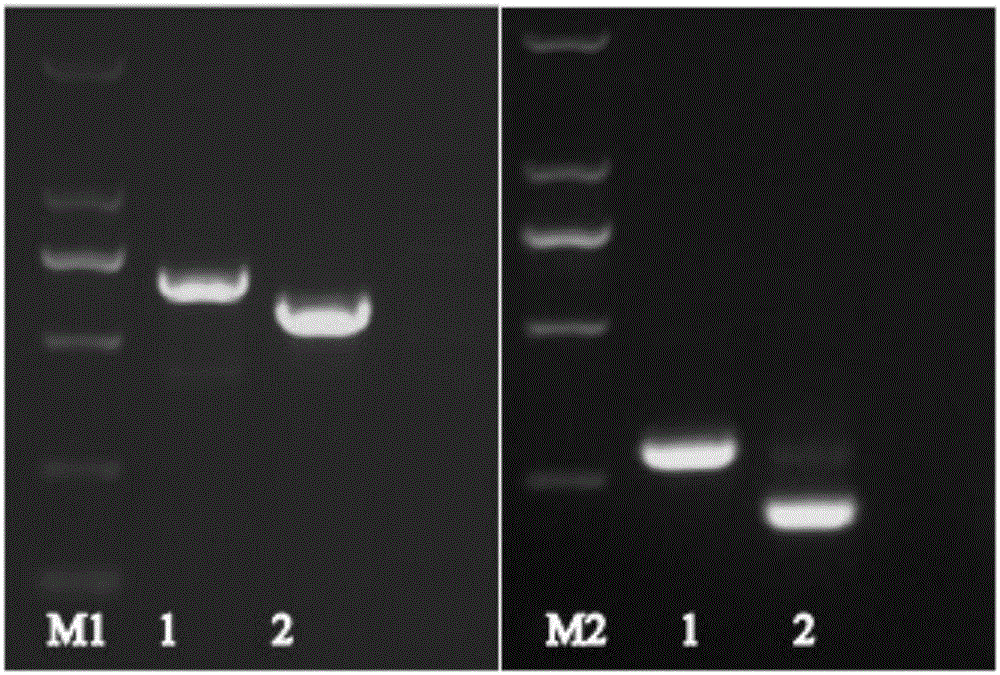
[0078] Dilute the ISKNV gradient to 10 0 、10 1 、10 2 、10 3 copy / mL, take 1mL of virus liquid to inoculate T25 cell culture flask, collect the cells at 7 days, 9 days and 11 days after inoculation respectively, prepare cDNA template according to the above method, and use the established Nest RT-PCR method for detection to determine the Methods To detect the sensitivity and minimum detection period of ISKNV mRNA, and set the cells not inoculated with virus as negative control. see results Figure 4 .
[0079] Figure 4 In M1.DNAMarker DL2000, the primers used are F3 and R3, 1. DNA positive template, 2. cDNA positive template, 3. Negative control, 4. Cell seeding 10 0 ~10 3 The cDNA samples obtained by incubation for 7 days with virus copy number;
[0080] The primers used by M2.DNAMarker DL2000 are F3, R3, 1. DNA positive template, 2. cDNA positive template, 3. Negative control, 4. Cell seeding 10 0 ~10 3 The cD...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com