Method for screening marker-free deletion mutant of Xanthomonas citri subsp.citri hfq gene
A technology of citrus canker and deletion mutants, which is applied in the field of molecular biology, can solve the problems of large workload, low efficiency, no marker-free deletion mutants of citrus canker hfq gene, etc., and achieves reduced workload and high screening efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
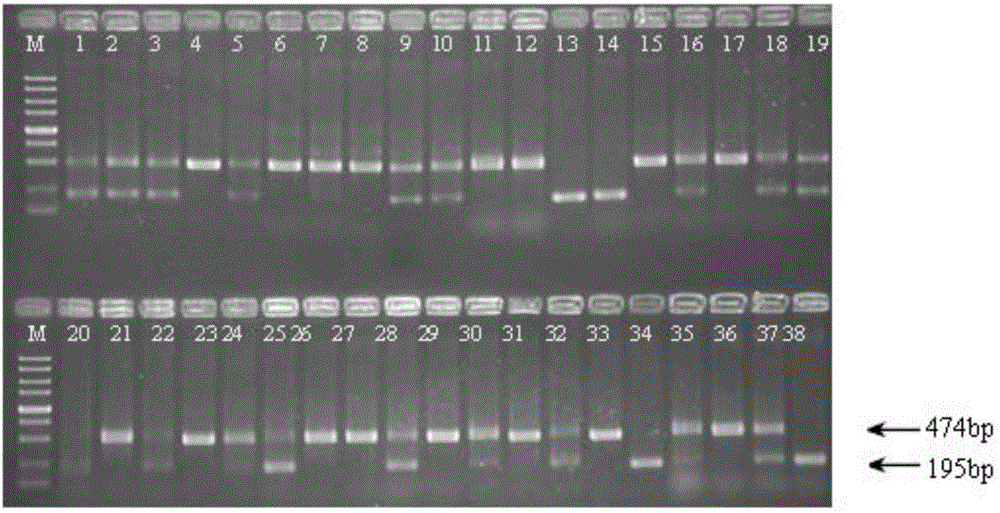
Image
Examples
Embodiment 1
[0043] Embodiment one, prepare experimental material
[0044] 1. The configuration of culture medium
[0045] The components of the liquid and solid medium used for E.coli cultivation are: Tryptone 10g / L, NaCl10g / L, Yeast Extract 5g / L, add water to dissolve, and finally adjust the volume to 1000mL, adjust the pH to 7.0-7.2, and pack After autoclaving. Add Agar 15g / L to LB solid medium.
[0046] The components of NA solid and NB liquid medium for Xanthomonas culture are: Polypeptone 5g / L, Sucrose 10g / L, Yeast Extract 1g / L, beef extract 3g / L, Agar 15g / L, dissolved in water , and finally set the volume to 1000mL, adjust the pH to 7.0-7.2, aliquot and autoclave, and remove Agar from the NB liquid medium.
[0047] 2. Main reagents and sources
[0048] Bacterial Genomic DNA Extraction Kit: Axygen, USA; Gel Recovery Kit, Plasmid Extraction Kit: Promega, USA; Southern Hybridization Kit: Roche, Germany; BamHI, KpnI, KpnI, XbaI, BMJM 109 Competent Cells, DH5α Large Intestine Bacill...
Embodiment 2
[0049] Example 2, Screening of X. citri hfq Gene Unmarked Deletion Mutants
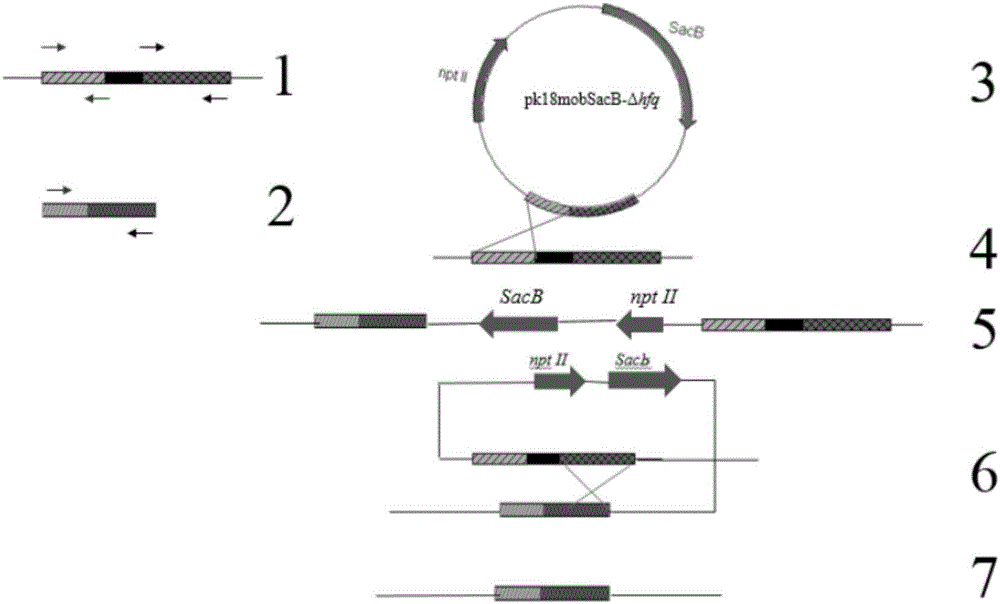
[0050] Its process is as follows figure 1 As shown, follow the steps below:
[0051] 1. Genomic DNA extraction of X. citri
[0052] The total DNA of X. citri bacterial culture was extracted by Axygen Bacterial Genomic DNA Extraction Kit.
[0053] 2. Construction of recombinant suicide plasmid vector with deletion of hfq gene
[0054] 1. Amplification of the upstream and downstream flanking sequences of the hfq gene of X. citri
[0055] (1) Amplify the flanking sequence upstream of the hfq gene of X. citri, and the amplification primers are:
[0056] Hfq-up-F: TG GGATCC CGCGTGTTGAAGGTGGTATT (SEQ ID No: 1, the underline is the BamH I restriction site)
[0057] Hfq-up-R: TG GGTACC CGAAAAATCCTCTTCATTATTGT (SEQ ID No: 2, the underline is the KpnI restriction site)
[0058] Using the previously extracted genomic DNA of X. citri as a template, use primers Hfq-up-F and Hfq-up-R to amplify the upst...
Embodiment 3
[0157] Embodiment 3, comparison of traditional and improved sucrose medium screening mutant methods
[0158] 1. The traditional sucrose medium screening steps for mutants are as follows:
[0159] (1) Pick a single colony screened by Kan+sugar-free NA after electroporation, and transfer it to 3 mL sucrose-free NB medium for overnight culture at 28°C and 200 rpm.
[0160] (2) Transfer the bacterial solution amplified in the previous step to 10% sucrose NB medium at a ratio of 1:100 at 28°C and 200rmp for 3-4 passages;
[0161] (3) Dilute the bacterium solution with OD600=0.5 after passage for 10 1 、10 2 、10 3 、10 4 、10 5 , pipette 60μL and spread on 10% sucrose NA plate, culture at 28℃ for 72h;
[0162] (4) Streak the single colonies on the 10% sucrose NA plate to Kan+sugar-free NA (with resistance) and 10% sucrose NA (without resistance) plates one by one, draw 100 single colonies, and culture at 28°C for 36h, Observe the growth of the colonies.
[0163] (5) Perform PCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com