Glucose dehydrogenase mutant, preparation method thereof and application
A technique for glucose dehydrogenase and mutants, which is applied in the field of glucose dehydrogenase mutants and its preparation, and can solve problems such as poor stability, difficult to reuse, and expensive coenzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The establishment of embodiment 1 genetically engineered bacteria
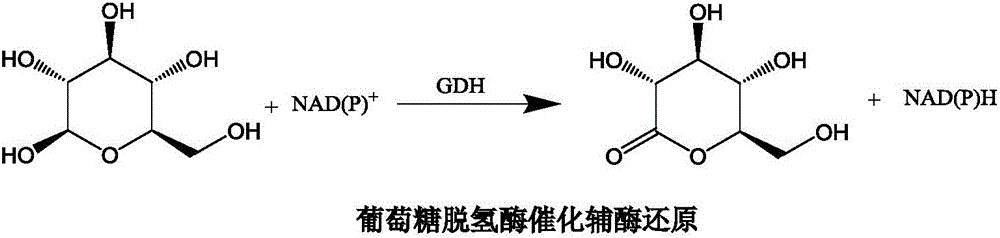
[0025] According to the glucose dehydrogenase gene GDH (GenBank: KU682779.1) included in NCBI, the glucose dehydrogenase gene fragment was artificially synthesized, and the gene fragment was used as a template to amplify and expand the fragment by PCR (adding NdeI and BamHI on both sides of the fragment) endonuclease fragment) its nucleotide sequence is shown in SEQ ID NO.3. And the gene was inserted into the pET-28a plasmid by using the NdeI and BamHI endonuclease sites, and the ligated vector was transferred into Escherichia coli BL21 (DE3) to establish the glucose dehydrogenase genetically engineered bacteria. The primers for PCR amplification of the glucose dehydrogenase gene are: the upstream primer is F1:5'-GGGCCATATGATGGACATGT ATCCGGATT-3' (SEQ ID NO.4), and the downstream primer is R1:5'-GCGGGGATCCTTAGCGGCC TGCCTGGAA T-3' (SEQ ID NO.5).
Embodiment 2
[0026] Embodiment 2 Obtaining of glucose dehydrogenase mutant gene
[0027] In this study, glucose dehydrogenase was protein engineered using an error-prone PCR method.
[0028] The 50 μl PCR reaction system is: 5 μl of 10× amplification buffer, 4 μl of each of the four dNTP mixtures (2.5 mmol / L), 50 pmol of each primer, 1.5 μg of template DNA, 0.5 μL of Taq DNA polymerase, Mg 2+ 2mmol / L, add double distilled water to 50μl.
[0029] The PCR amplification program was: pre-denaturation at 95°C for 3 min, denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, and 30 cycles; further extension at 72°C for 5 min, cooling to 4°C, and ending the reaction.
[0030] experiment process
[0031] Chemically synthesize the glucose dehydrogenase gene and insert the gene into the pET-28a plasmid using NdeI and BamHI endonuclease sites as a template for gene mutation; error-prone PCR amplifies the glucose dehydrogenase gene, and the amplified gene fragments a...
Embodiment 3
[0034] The shaking flask culture of embodiment 3 recombinant escherichia coli
[0035] The recombinant Escherichia coli obtained in Example 1 and Example 2 were inoculated into LB medium (10 g / L of peptone, 5 g / L of yeast extract, 10 g / L of NaCl) containing 5 mL of kanamycin (50 μg / mL) respectively. , pH7.0), at 37°C, shake culture at 200rpm for 4-8 hours. Transfer 1 mL of bacterial culture solution to 50 mL of LB liquid medium (containing 50 μg / mL of kanamycin), shake culture at 37°C and 200 rpm until the OD600 is 0.6-0.8, and then add the inducer IPTG to the final concentration After inducing expression at 22-26°C and 200rpm for 8-12 hours, the cells were collected by centrifugation (8000rpm, 15min, 4°C) and washed twice with phosphate buffer (pH7.5, 10mmol / L). Disperse in the same pre-cooled buffer and perform sonication in an ice-water bath. Centrifuge (10000rpm, 15min, 4°C), discard the cell fragments, and obtain glucose dehydrogenase and its mutant crude enzyme solutio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com