Gene saturation mutation library and its construction method and application
A construction method and gene technology, applied in the field of gene saturation mutation library and its construction, can solve the problems of cumbersome steps, low mutation efficiency of multiple bases, inability to achieve target gene sequence detection, etc., and achieve the effect of high mutation success rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
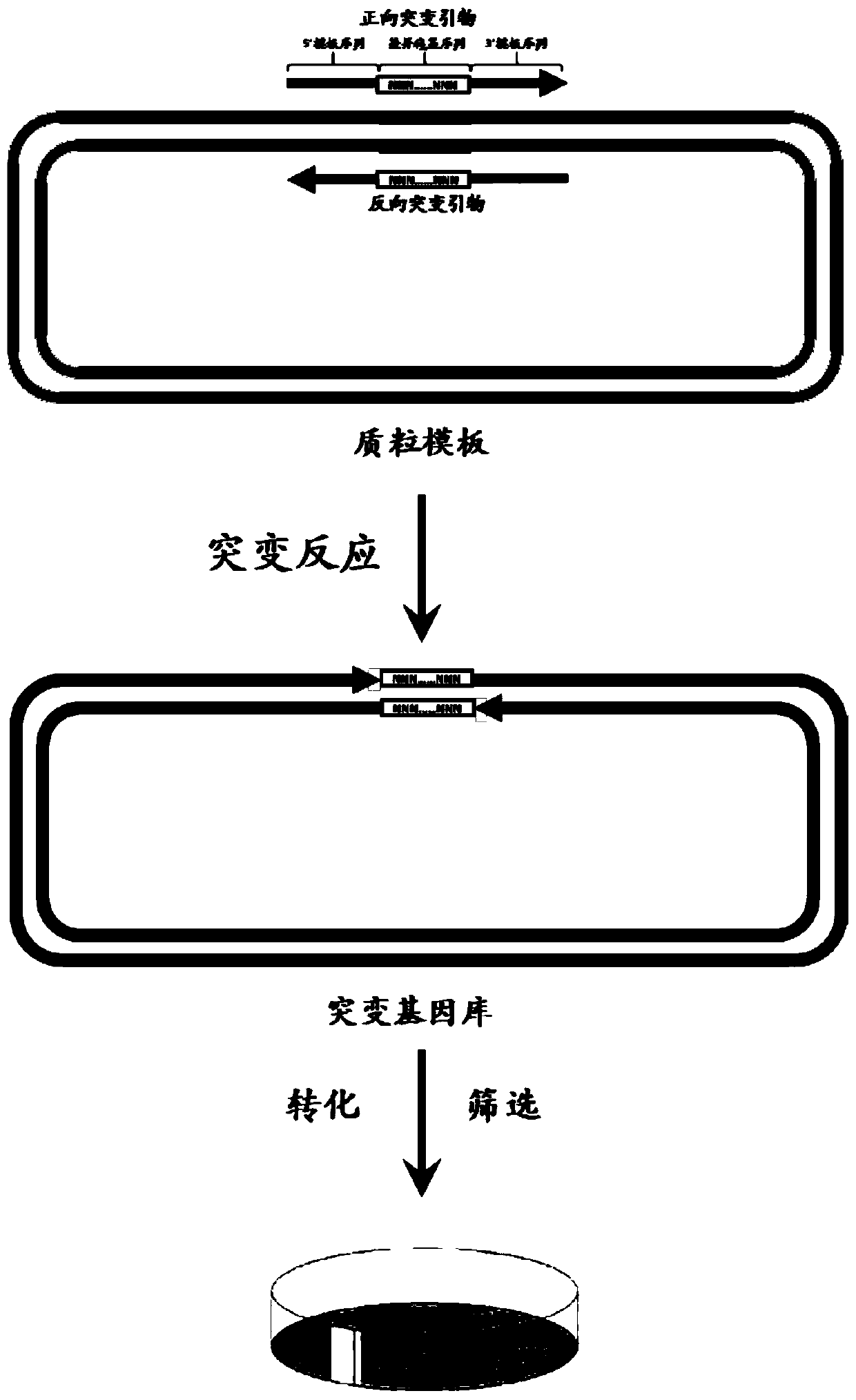
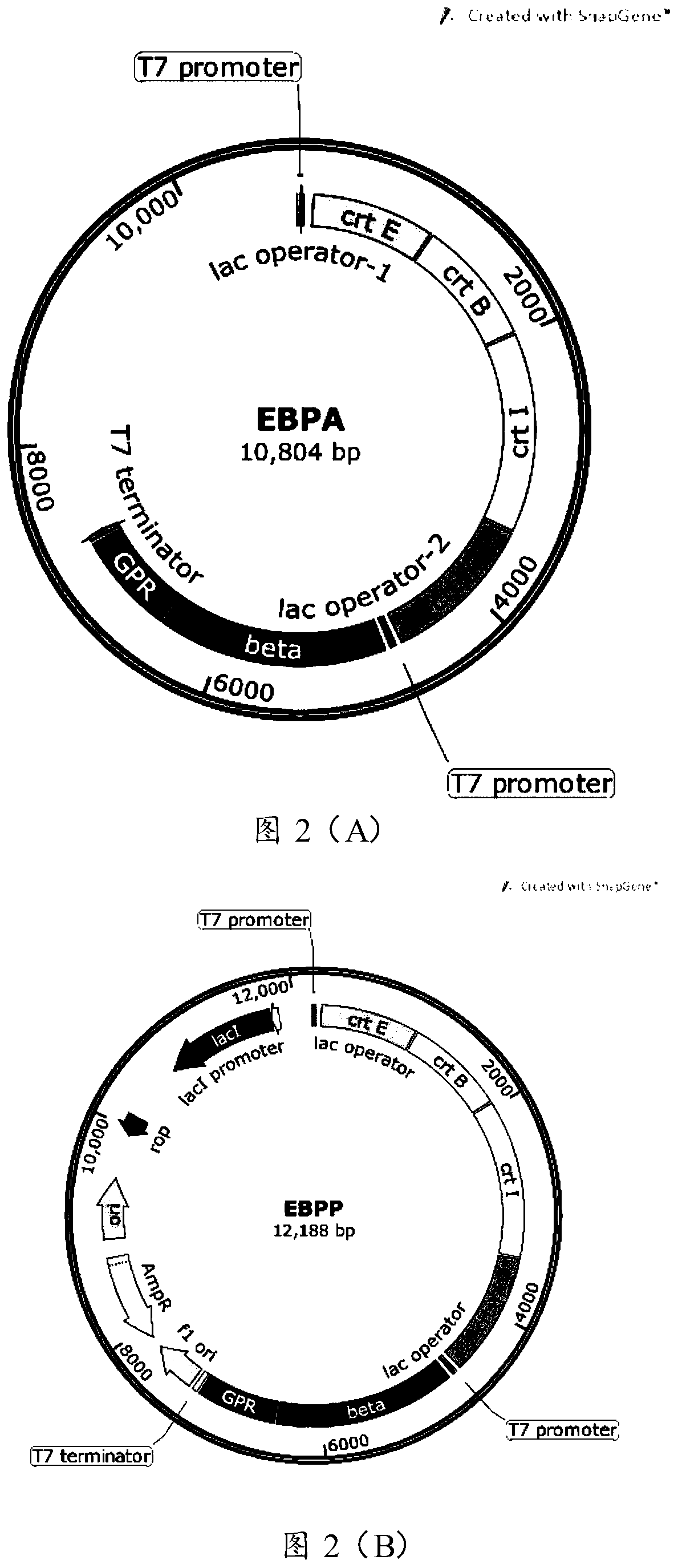
[0054] The natural active PR can absorb green light in a certain wavelength range due to the combination of trans-retinal, so that the bacteria containing it have different degrees of red or orange. 5 genes related to the PR gene (750bp, sequence shown in SEQ ID No.6) and retinal synthesis-crtE (909bp, sequence shown in SEQ ID No.1), crtB (930bp, sequence shown in SEQ ID No.1) ID No.2), crtI (1479bp, sequence as shown in SEQ ID No.3), crtY (1149bp, sequence as shown in SEQ ID No.4) (Genbank: 90087) and β-diox (1701bp, sequence as shown in Shown in SEQ ID No.5, Genbank: AF271298) were all constructed into the pACYCDuet-1 vector (4008bp, purchased from Merck Company), and the recombinant plasmid EBPA with a total length of 10.804kb was obtained (spectrum as shown in figure 2 (A) shown). Such as image 3 As shown, the DNA sequence of the target gene PR was divided into 62 groups with each 12bp as a group, and the EBPA recombinant plasmid was used as a template to design satura...
Embodiment 2
[0056] PR gene (750bp) and 5 genes related to retinal synthesis - crtE (909bp), crtB (930bp), crtI (1479bp), crtY (1149bp) (Genbank: 90087) and β-diox (1701bp, Genbank: AF271298) were all constructed into the pETDuet-1 vector (5420bp, purchased from Merck Company), and the recombinant plasmid EBPP with a total length of 12.188kb was obtained (the map is shown in figure 2 (B) shown). Using the EBPP recombinant plasmid as a template, the image 3 The numbers in the PR sequence are 6, 7, 8, 9, 12, 13, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 27, 28, 31, 32, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 47, 48, 51, 52, 53, 54, 55, 56 sequences (40 groups in total) design saturation mutation primers for saturation mutation Library construction.
[0057] The sequences of the mutation primers are shown in Table 1:
[0058] Table 1
[0059]
[0060]
[0061]
[0062]
[0063]
[0064] The reaction system was EBPP plasmid 80ng, forward mutation primer 150nM, rever...
Embodiment 3
[0068] Example 1 application of the number prepared by the present invention is 26, 29, 57 and 58 and embodiment 2 application of the number prepared by the present invention is 9, 15, 19, 24, 27, 45, 48 and 55 of each group of overnight culture The number of clones on the plate was more than 500, and 20 clones were randomly selected for sequencing. The mutation success rate is shown in Table 2. Among them, the "number of effective sequencing samples" refers to the total number of samples with successful sequencing reactions among the 20 samples sent for testing. "Number of mutants" refers to the total number of different mutant sequences obtained in the samples with successful sequencing reactions. If multiple samples have the same mutant sequence, only one sequence is recorded. The average mutation rate of EBPA is 76.6%, and EBPP The average mutation rate was 85.8%. Since the number of clones grown on the plates cultured overnight in the control group was less than 50, for t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com